Introduction
Deciphering the biological impacts of millions of single nucleotide variants remains a major challenge. Recent studies suggest that RNA modifications play versatile roles in essential biological mechanisms, and are closely related to the progression of various diseases including multiple cancers. To comprehensively unveil the association between disease-associated variants and their epitranscriptome disturbance, we built RMDisease, a database of genetic variants that can affect RNA modifications. By integrating the prediction results of 18 different RNA modification prediction tools and also 303,426 experimentally-validated RNA modification sites, RMDisease identified a total of 202,307 human SNPs that may affect (add or remove) sites of eight types of RNA modifications (m6A, m5C, m1A, m5U, Ψ, m6Am, m7G and Nm). These include 4,289 disease-associated variants that may imply disease pathogenesis functioning at the epitranscriptome layer. These SNPs were further annotated with essential information such as post-transcriptional regulations (sites for miRNA binding, interaction with RNA-binding proteins and alternative splicing) revealing putative regulatory circuits. A convenient graphical user interface was constructed to support the query, exploration and download of the relevant information. RMDisease should make a useful resource for studying the epitranscriptome impact of genetic variants via multiple RNA modifications with emphasis on their potential disease relevance.
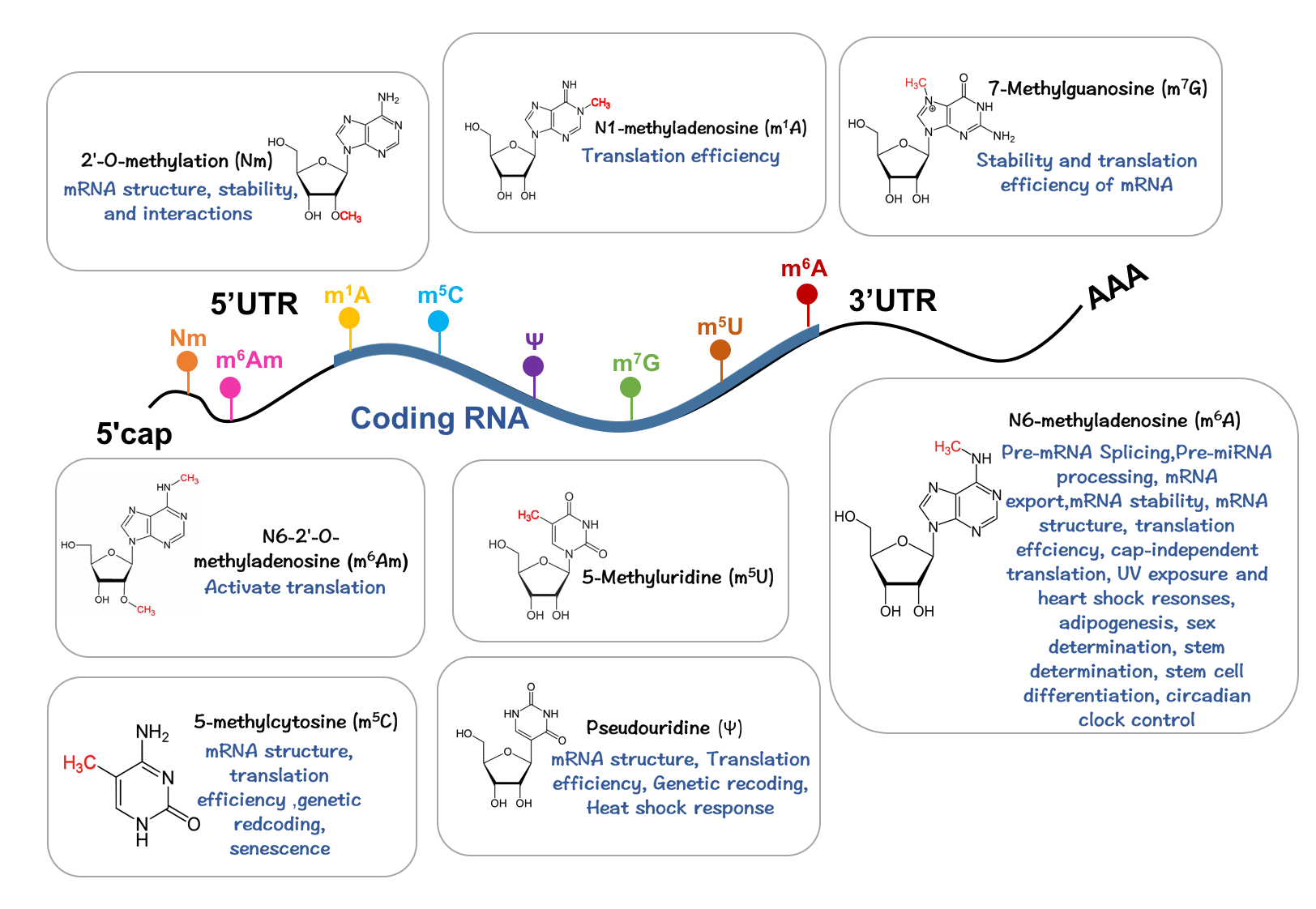
8 type modifications
We consider in this study only the RNA modifications that widely occur in the transcriptome. Since there is yet available a relatively complete high-confidence collection of such data, we manually collected from 33 studies (86 high-throughput sequencing samples) the sites of eight types of RNA modifications, including m6A (178,049 sites), m5C (95,391 sites), m1A (16,346 sites), m5U (3,696 sites), Ψ (3,137 sites), m6Am (2,447 sites), m7G (2,525 sites) and Nm (1835 sites), respectively. These sites were reported from 86 samples generated by 18 base-resolution technologies, including m6A-REF-seq, MAZTER-seq, miCLIP, m6A-CLIP-seq, PA-m6A-seq, Ψ-seq, Pseudo-seq, CeU-Seq, RBS-Seq, m1A-MAP, m1A-seq, Aza-IP, RNA-BisSeq, FICC-Seq, Nm-seq, m7G-seq, m7G-miCLIP-Seq.
 [1] Li, Xiaoyu, Xushen Xiong, and Chengqi Yi. "Epitranscriptome sequencing technologies: decoding RNA modifications." Nature methods 14.1 (2017): 23-31.
[1] Li, Xiaoyu, Xushen Xiong, and Chengqi Yi. "Epitranscriptome sequencing technologies: decoding RNA modifications." Nature methods 14.1 (2017): 23-31.
| New Nomenclature |
Name | Short Name | RNAMods abbrev. | Formula | Monisotopic Mass | Average Mass |
|---|---|---|---|---|---|---|
| 6A | N6-methyladenosine | m6A | = | C11O4N5H15 | 281.1124 | 281.2719 |
| 06A | N6,2'-O-dimethyladenosine | m6Am | X | C12O4N5H17 | 295.1281 | 295.2928 |
| 1A | 1-methyladenosine | m1A | " | C11O4N5H15 | 281.1124 | 281.2719 |
| 5C | 5-methylcytidine | m5C | ? | C10O5N3H15 | 257.1012 | 257.2467 |
| 5U | 5-Methyluridine | m5U | T | C10O6N2H14 | 258.0582 | 258.0852 |
| 9U | Pseudouridine | P(Ψ) | Y | C9O6N2H12 | 244.0695 | 244.2043 |
| 0A | 2'-O-methyladenosine | Am | : | C11O4N5H15 | 281.1124 | 281.2719 |
| 0C | 2'-O-methylcytidine | Cm | B | C10O5N3H15 | 257.1012 | 257.2467 |
| 7G | 7-methylguanosine | m7G | 7 | C11O5N5H17 | 299.123 | 299.2867 |
| 0G | 2'-O-methylguanosine | Gm | # | C11O5N5H15 | 297.1073 | 297.2712 |
| 0U | 2'-O-methyluridine | Um | J | C10O6N2H14 | 258.0852 | 258.2313 |
User Guide
RMDisease provides users with a browser page for 8 types of RNA modifications (m6A, m5C, m1A, m5U, Ψ, m6Am, m7G, and Nm). The detailed list of modified sites information is listed here, according to the needed filter selection of modification types, RNA types, RNP,miRNA Target, SplicingSite, GWAS and ClinVar.
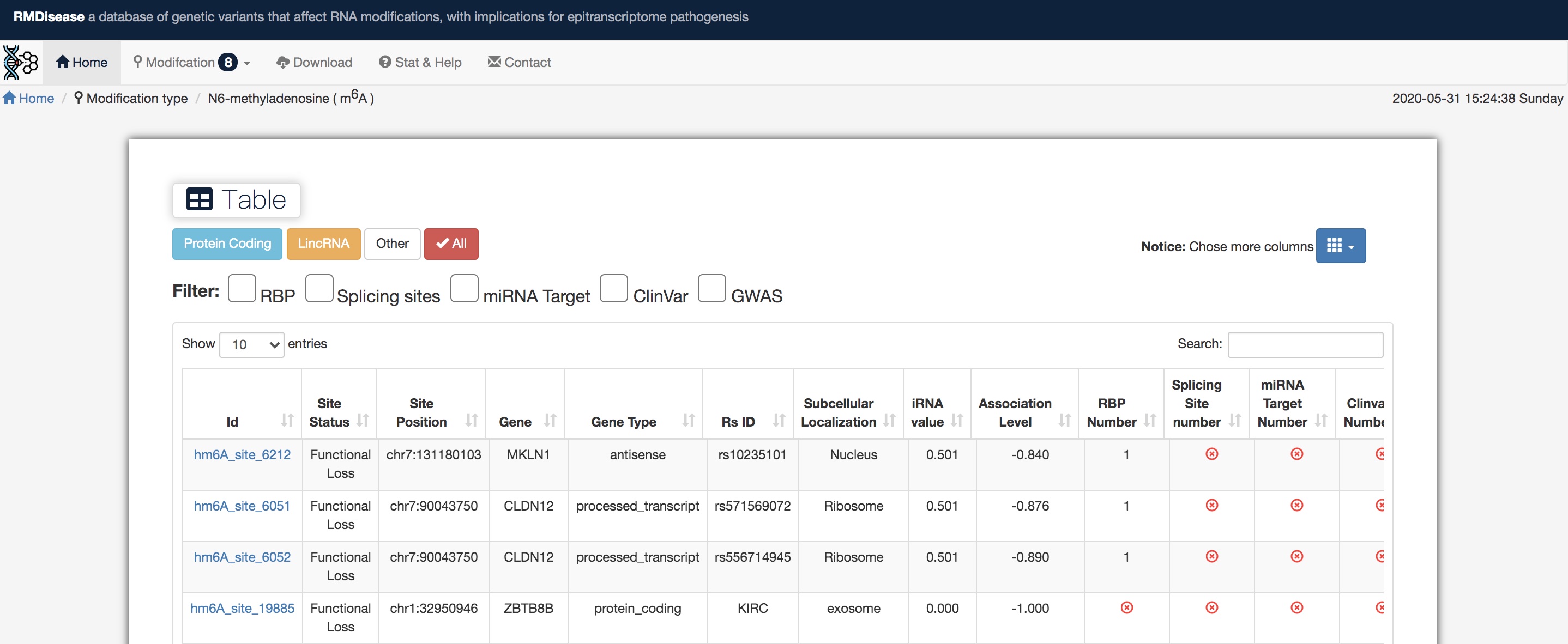
Click on button, after turning the scroll bar to the right, to link to the Jbrowser showing SNP-sites, genes, and different modification sites context.
#Search
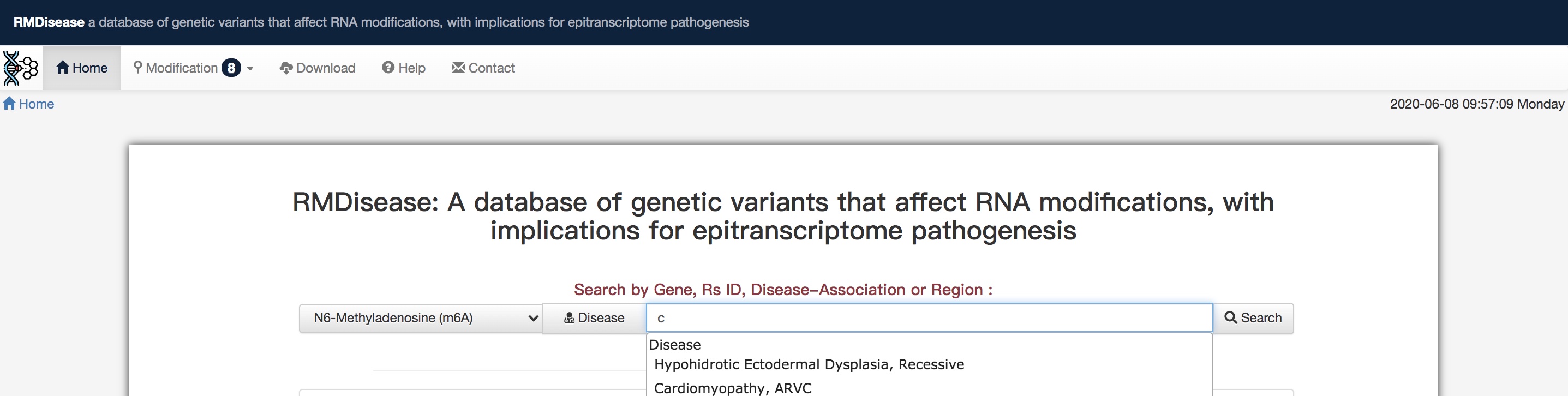
>> RMDisease provides users a combined selection function of several choices to query the various data which you are interested in. The first choice is to select the interesting modifications of interest, e.g.i.e. m6A. The second choice is used to determine the searching approach, i.e. searching depending on Gene, Rs ID, Disease, or Region.
√Searching disease
For example, after setting the Disease choice, users can type in any letters to trigger the drop-down box correspondingly, and then select the specific diseasegene name which you want (e.g. typing in cancer and selecting the specific cancer disease name). Or you could direcly type in cancer and Press the button,. The query results will provide you all types of cancer diseases associated information in RMDisease. Hhere you go!
√Explain the searching disease results
>> The detailed list of search results containing disease site-associated m6A sites, corresponding annotation and disease information.
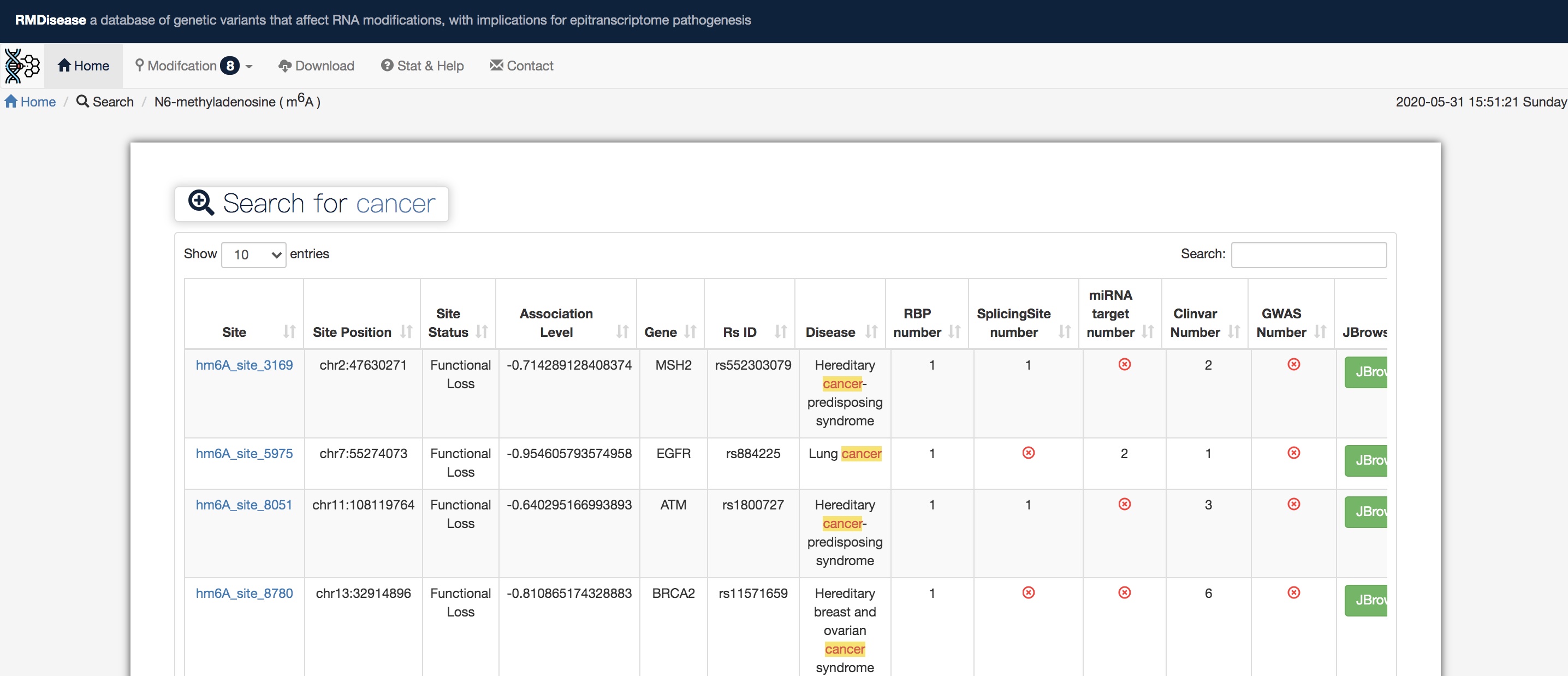
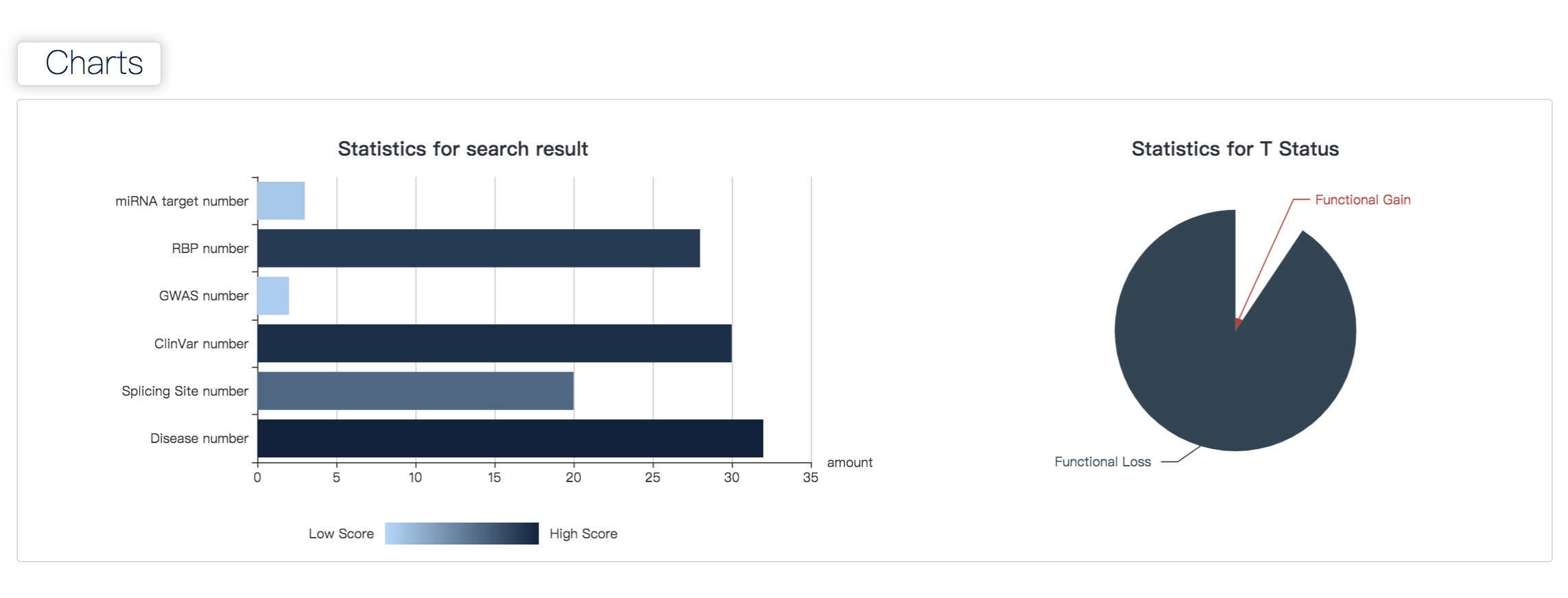
>> Statistic bar and pie graphs illustrate thefor sources of detected disease-associated sites, including Statistics of annotation information (miRNA Target, RBP, GWAS, ClinVar, Splicing Site and Disease) and T Status (Functional Gain or Functional Loss). In this case, we finding 32 m6A sites associated with cancer records, 29 of whichthem are Functional Gain and 3 are Functional Loss in T status.
√Explain the details of an individual searching result
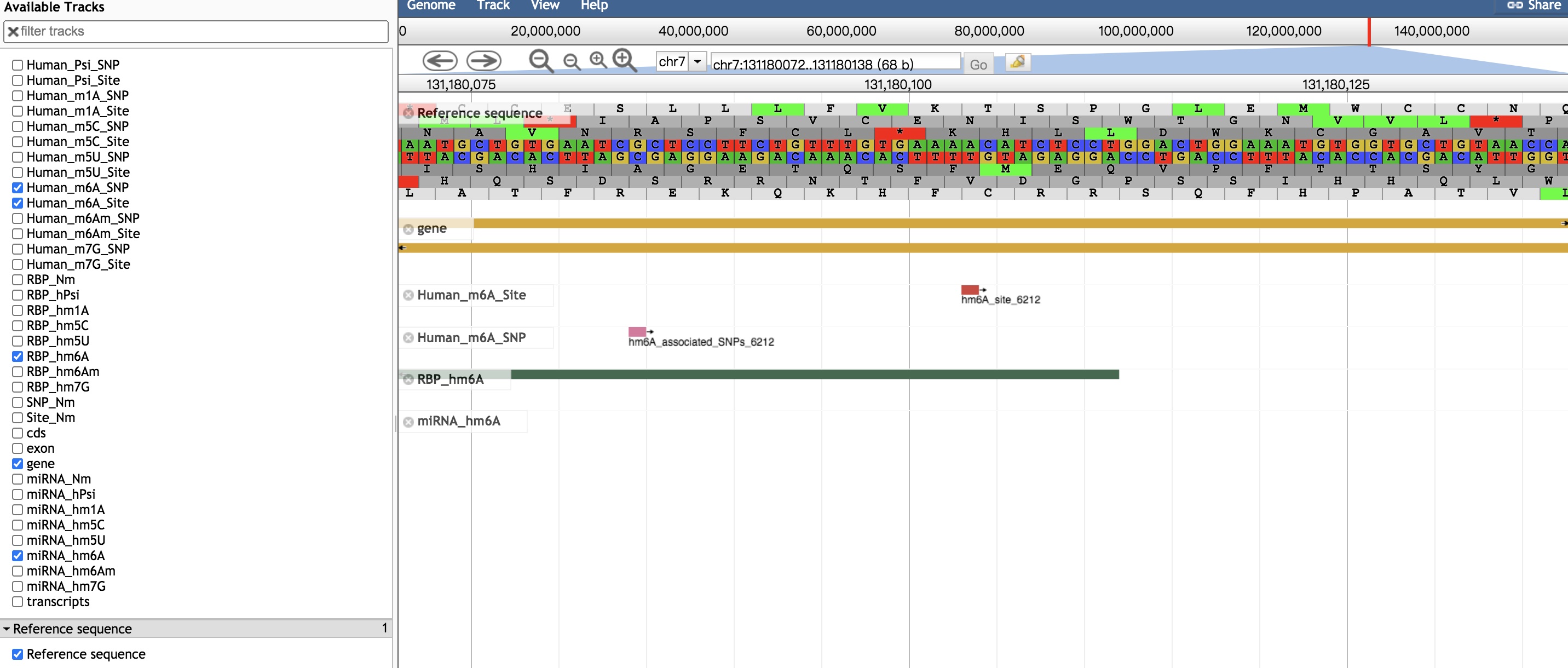
>> Click on button, after turning the scroll bar to the right, users can explore more details of a modification site.
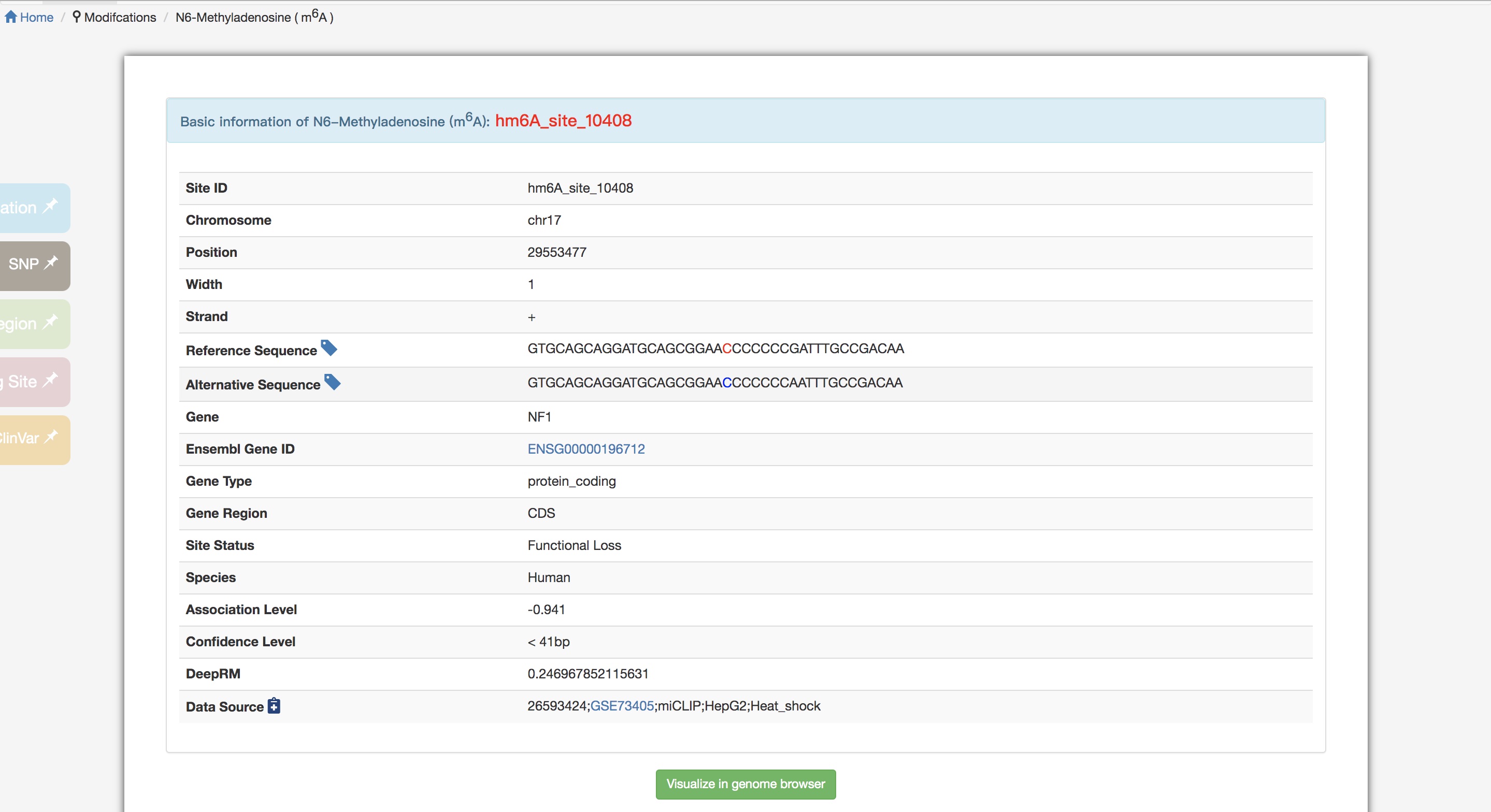
>> Basic information about the modification site. Hover icon to know the explanation for its relevant concept. It is noted that within both the reference and alternative sequences, the blue nucleotide represents the variant, and the red one is the corresponding modification site. Click to link to Jbrowse to show modification sites, SNP sites, miRNA target and RBP Binding sites context.
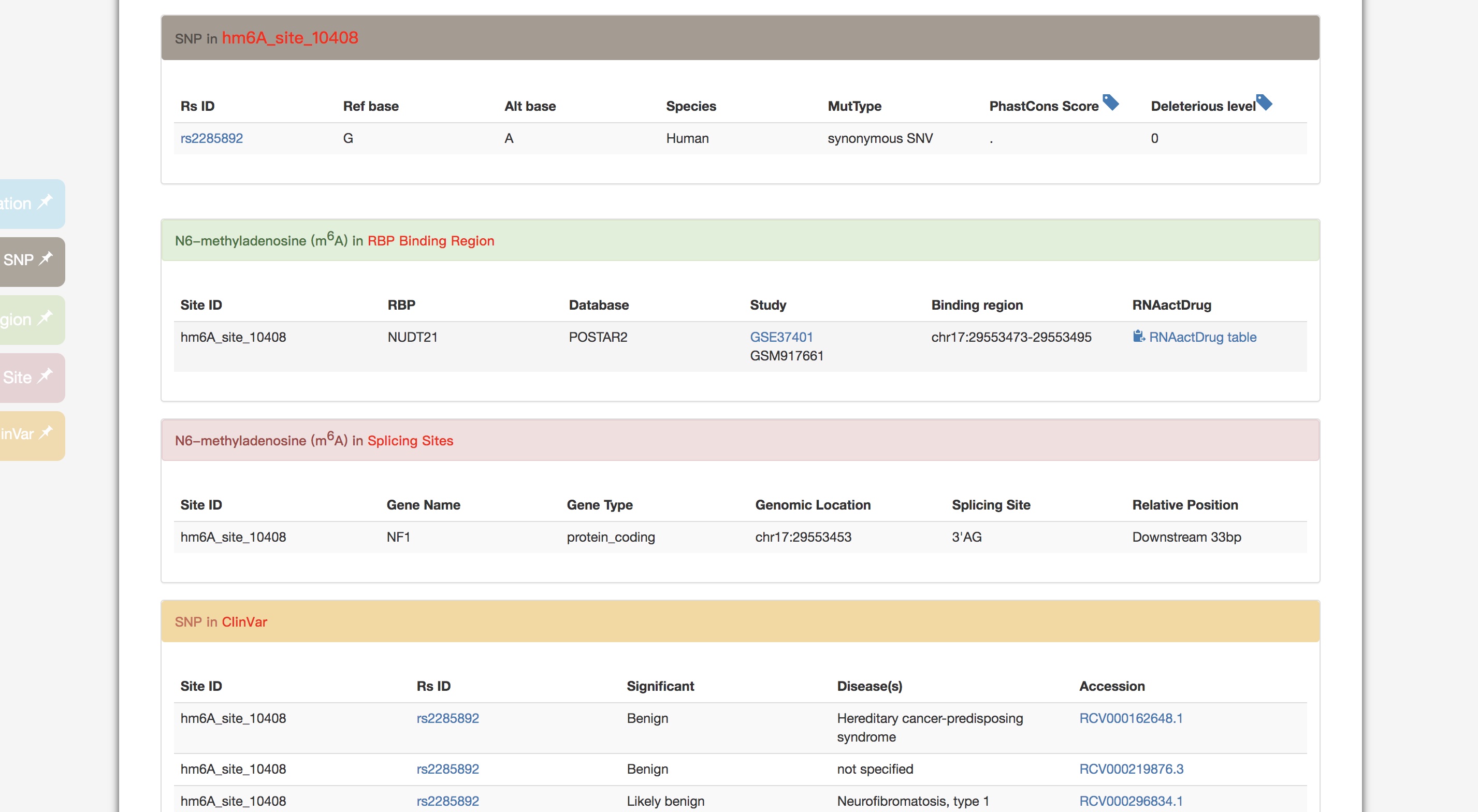
>> More details about m6A variants as well as the related RNA binding region, miRNA targets, splicing sites and diseases information.
# Download
>> The used data on this website from RMDisease is available.
- MODOMICS
MODOMICS is a database of RNA modifications that provides comprehensive information concerning the chemical structures of modified ribonucleosides, their biosynthetic pathways, the location of modified residues in RNA sequences, and RNA-modifying enzymes. - Met-DB v2.0
Met-DB v2.0, the significantly improved second version of Met-DB, which is entirely redesigned to focus more on elucidating context-specific m6A functions. Met-DB v2.0 has a major increase in context-specific m6A peaks and single-base sites predicted from 185 samples for 7 species from 26 independent studies. - RMBase v2.0
RMBase v2.0 is a comprehensive database that integrates epitranscriptome sequencing data for the exploration of post-transcriptional modifications of RNAs and their relationships with miRNA binding events, disease-related single-nucleotide polymorphisms (SNPs) and RNA-binding proteins (RBPs). - CVm6A
CVm6A identified 340,950 and 179,201 m6A peaks from 23 human and eight mouse cell lines respectively, and classified them according to subcellular components, gene regions and relevance to cancers. - REPIC
The REPIC (RNA Epitranscriptome Collection) database records about 10 million peaks called from publicly available m6A-seq and MeRIP-seq data using our unified pipeline. These data were collected from 672 samples of 49 studies, covering 61 cell lines or tissues in 11 organisms. - RNAmod
RNAmod is an interactive, one-stop, web-based platform for the automated analysis, annotation, and visualization of mRNA modifications in 21 species. . - m6AVar
m6AVar is a comprehensive database of m6A-associated variants that potentially influence m6A modification, which will help to interpret variants by m6A function.