Introduction of m6AmPred
N6,2’-O-dimethyladenosine (m6Am) is a reversible modification widely occurred on varied RNA molecules. The biological function of m6Am is yet to be known though recent studies have revealed its influences in cellular mRNA fate. Precise identification of m6Am sites on RNA is vital for the understanding of its biological functions. We present here m6AmPred, the first web server for in silico identification of m6Am sites from the primary sequences of RNA. Built upon the eXtreme Gradient Boosting with Dart algorithm (XgbDart) and EIIP-PseEIIP encoding scheme, m6AmPred achieved promising prediction performance with the AUCs greater than 0.954 by 10-fold cross-validation and independent testing datasets. To critically test and validate the performance of m6AmPred, the experimentally verified m6Am sites from two data sources were cross-validated. An average AUC of 0.902 under mature mRNA mode was achieved, showing reasonable accuracy and reliability of m6AmPred. We believe that our model should make a useful tool for the researchers who are interested in N6,2’-O-dimethyladenosine RNA modification.
m6AmPred takes standard FASTA format as input file.
User should provide as least one 81bp RNA sequence centered on an A base.
Example:- >test1
AUGGGGGUGGAACUCAUGAUGGAAUUGGAGCCUUUACAAGGGAAUGAAGA GACAAGAGCUCUCUUUAUGCCACGUGAGGAUACAGCAAGGCCCCAAUCUG CAAGCCAGGAAGAGUCGUCACGAGAACCAGACCAUGCAGGAACUCUGAUC
GUGGA
(1) Chose the model type (Full transcript or Mature mRNA mode) to start the analysis, users can get notification email when the job is finished.
(2) When users submit their job, an unique job ID will be created. Users can view and download their prediction results when the job is finished.
(1) Firstly, a map of all putative m6Am sites in a given sequence is provided, users can know the position of each m6Am modification clearly.
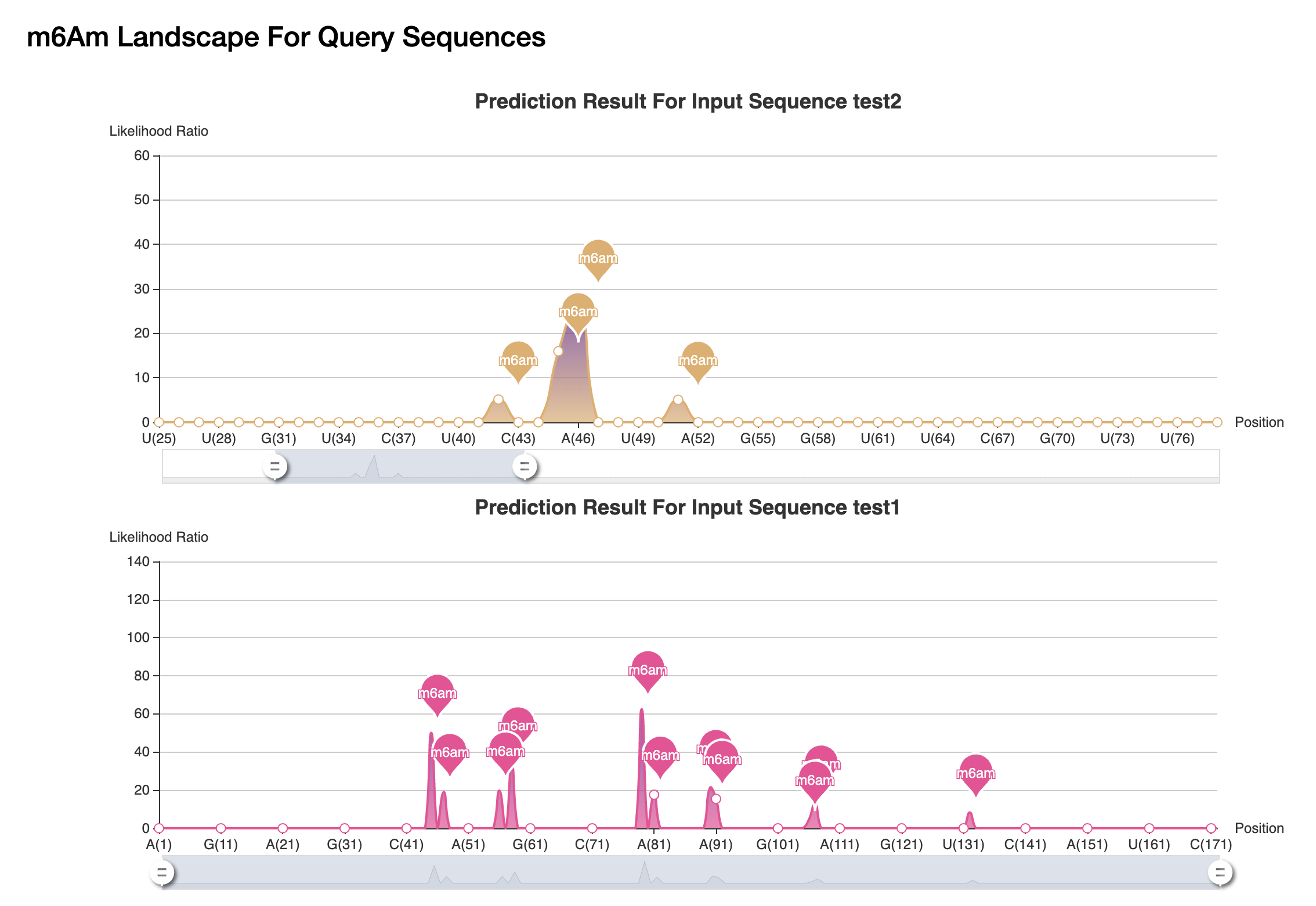
(2) The detail information of each computational predicted m6Am site are then listed, with m6Am position, sample name, likelihood ratio, and decision. The detailed explanation for each column is provided as well.