m6A-Maize Help Document
We present here m6A-Maize, a centralized online platform for predicting the m6A methylation degree of mRNA and the SNP-related traits in Maize. The m6A-Maize consists of the following two major components, including:
1) Sequence Predictor: a web server for predicting the m6A modification possibility in Maize using Weakly Supervised Model.
2) SNP Predictor: a web server for predicting the SNP with m6A modification and obtain the report of related traits information for download.
3) Database: a database for all SNPs data information in our research, which contains 3 data tables that have been classified according to confidence level and negative or positive mutation.
Sequence Predictor
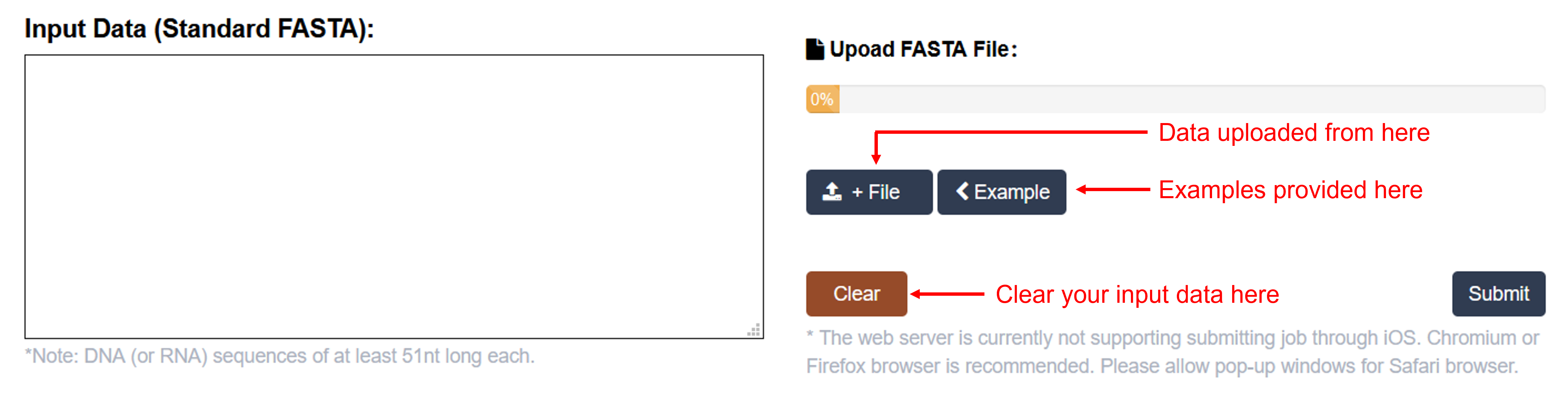
Data input
m6A-Maize Sequence Predictor accepts FASTA format.
(1): The standard FASTA format.
Example:
>example1
GGCATCCGTTACCGCCCATCACCCTCGGAGCCTCCATCAACGAGCACCAAGA
AAGGGAAGGGAGCTGCCTCAGGCCCTACGCAGTTCGTCCACACGCTCAACG
CTACAGCAGTGGCGGTGCCTCGTCTGATAGTTTGCATTCTAGAGAATTTTCAG
CAGGGCGATGGGTCGATTGTGGTTCCAGAGCCACTAAGACCCTATATGGGTG
GACTAGAGTTGCTCTCCCCAAAATTCAAGTGACCCTCTACGGTCACCCGCTG
TTGTGCTTGTGTGGACTTAGAACTGTCTGGCTGAATACAAGTTAAAG
>example2
ATATGCCATGGATAATGCACGCGGGAACGGAACAAAGACACGCGGCGTGCG
GGGCCTCGCTCCTCTGGTCTTCCCTCCAGCCCTCGACGGTGGTCATGGCCG
CCGCCGCCGCCACTTTCGGCTTCCTCCATCCTCC
>example3
TCGCTGCTATTGATGTGTCTTGTGTTGGTAGACTAAAGGATTTAAGAGCCATGT
GTACAAAATTCAGTTTACAAGCAAAGATCAAAGGTGGATGGATGTTTGATATGC
ATACGAAGTCTTCCTTTT
>example4
ATGGGGAGGCTCGGTTTTAGGAGGAAGGTGTATCTCATGGGACGGAGCAAG
CCTGACCACGGCGTGGTGCCGCTCCTCGAGAGCTTGGGAATGCGTCTCTCC
TCGGCCAAACTCATCGCGCCTTACGTCGCGGCTGCGGGCCTTACTGTGCTGA
TTGATAGGGT
(2): Users can either use the example file we provided or upload their own file.
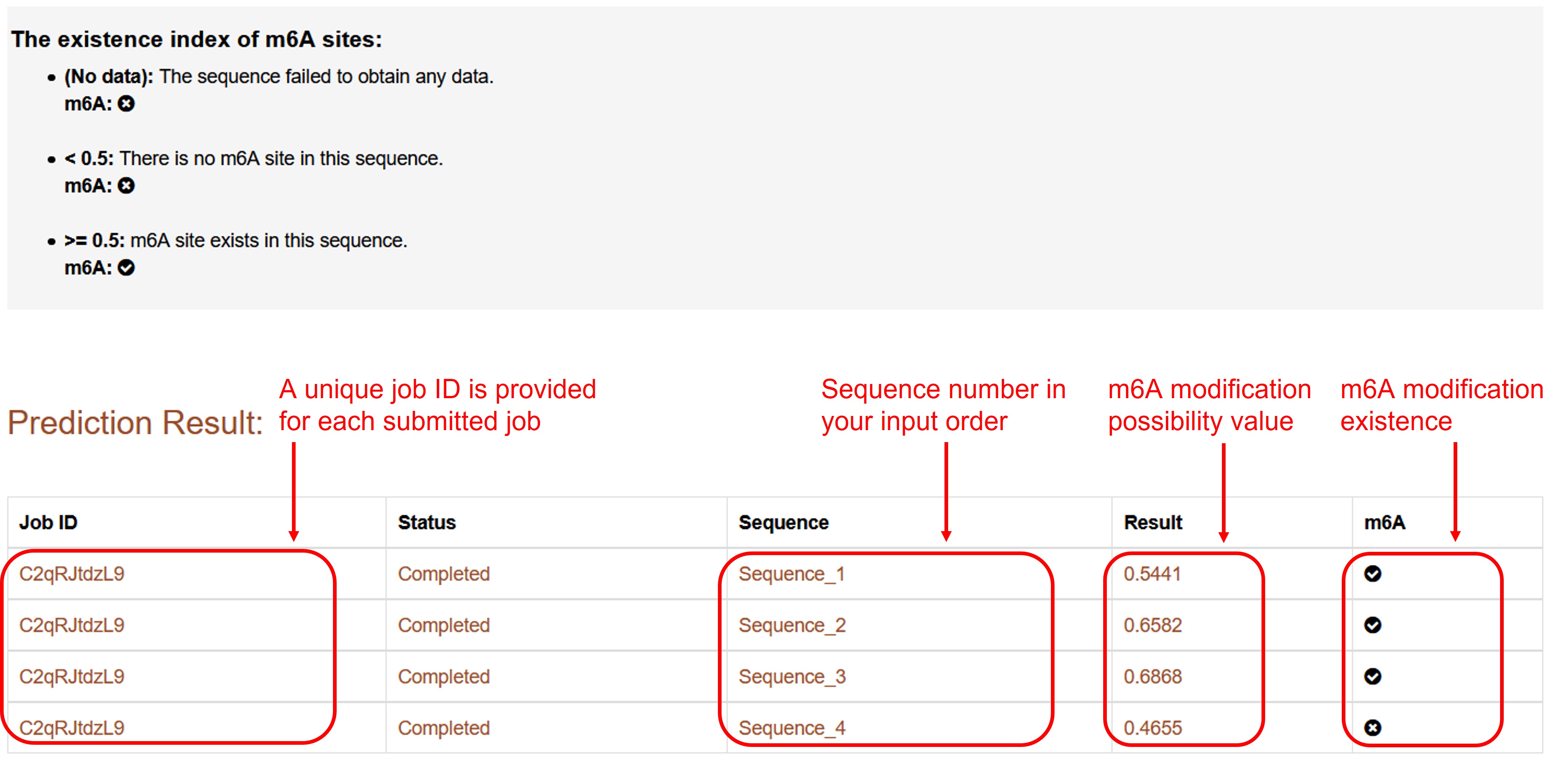
Get results
The m6A sites prediction result of the predictor will be displayed on the result page.
SNP Predictor
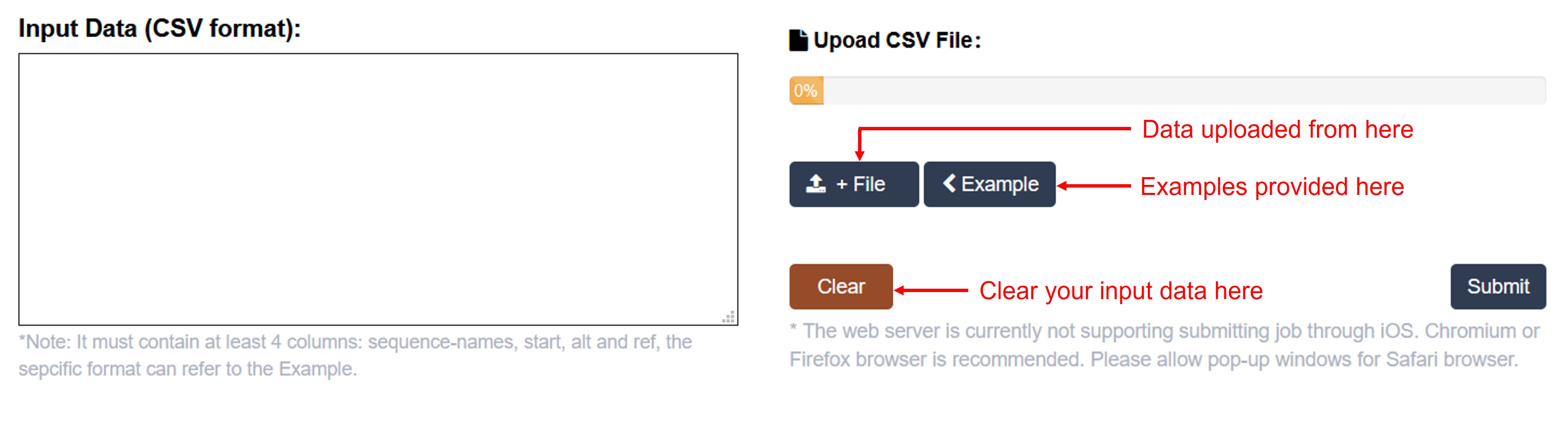
Data input
m6A-Maize SNP Predictor accepts CSV format.
(1): The tab-delimited format, which contains SNPs information with four columns: chromosome, start position, mutation base, and reference base. The header must be writen on first line and each column should be delimited by a comma.
Example:
seqnames,start,alt,ref
10,104454,T,C
10,26774840,T,G
1,148688773,C,T
1,2977148,T,A
9,157739331,G,A
1,4894903,G,C
8,4551689,C,T
9,157487101,G,A
1,16415903,A,G
(2): Users can either use the example file we provided or upload their own file.
Get SNP results
The SNP-related traits prediction result of the predictor will be displayed on the result page.
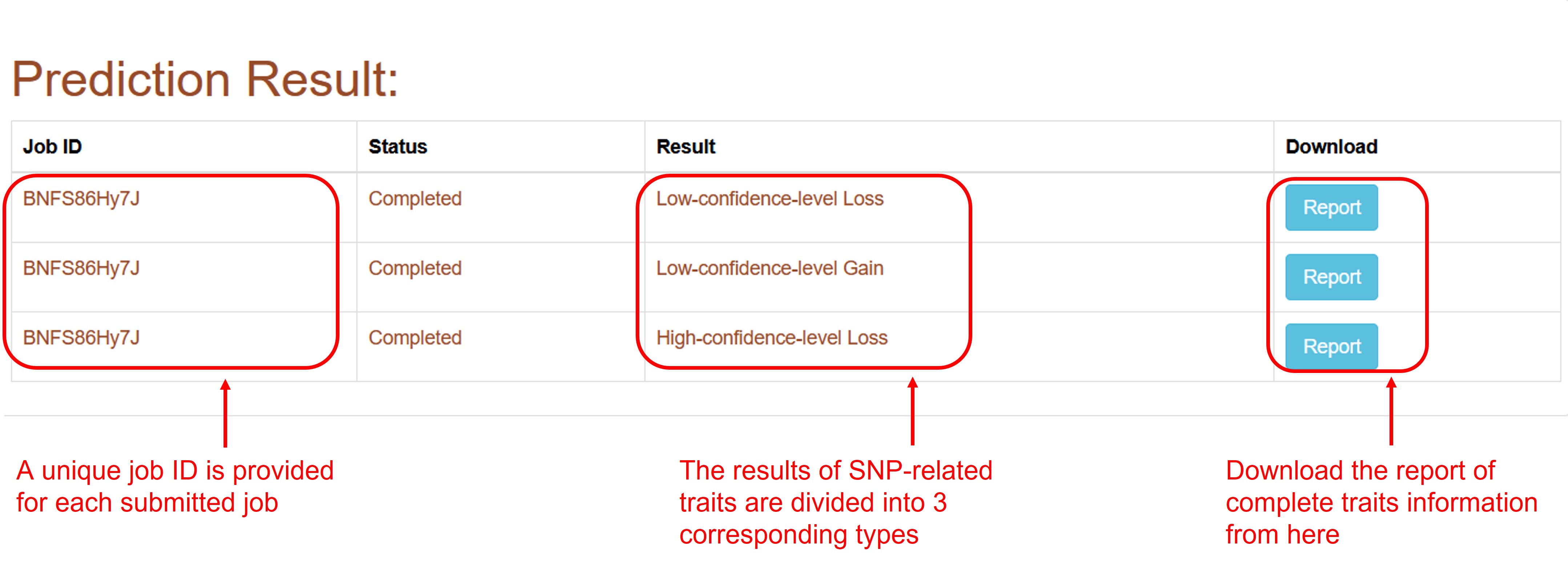
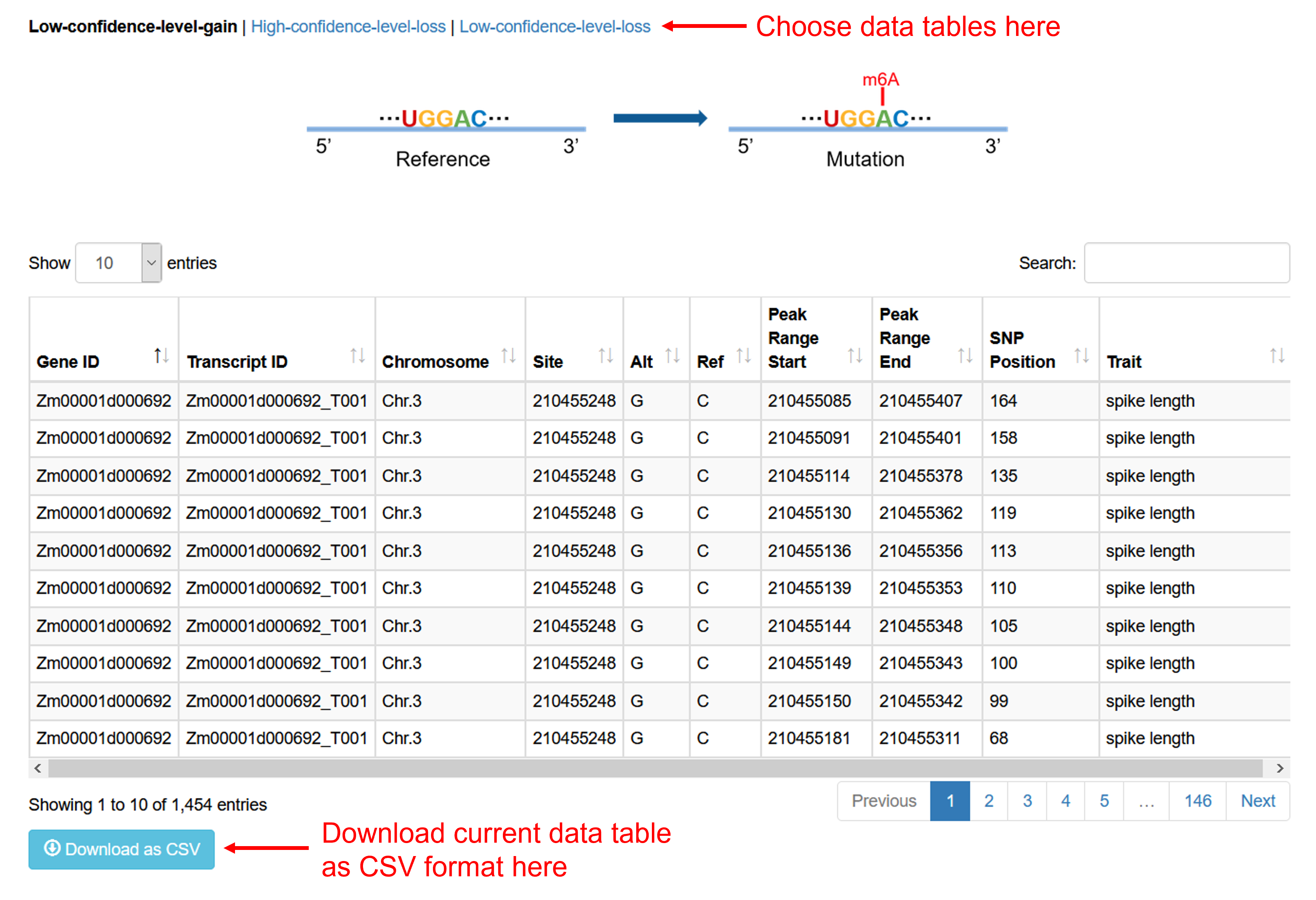
Database
For m6A-Maize Database, all SNPs data of m6A modification in Maize are provided. Users can query and download SNP information related to GWAS database. In addition, the m6A SNP information have been classified according to confidence level and negative or positive mutation and display in 3 different tables, you can download them as CSV file for each data table.
(1): Low-confidence-level-gain
The original sequence has not been verified by MeRIP-seq, but the model predicts that there may be no m6A modification, and the mutated sequence is predicted to have an increased degree of m6A modification. This is considered low-confidence-level-gain.
(2): Low-confidence-level-gain
The original sequence was verified by MeRIP-seq to contain m6A modification, and the mutated sequence was predicted by the model to reduce the degree of modification. This is called loss. At the same time, we think this is high-confidence.
(3): Low-confidence-level-gain
The original sequence has not been verified by MeRIP-seq, but the model predicts that there may be m6A modification, and the mutated sequence is predicted to have a reduced degree of m6A modification. This is considered low-confidence-level-loss.