What is a ConsRM score
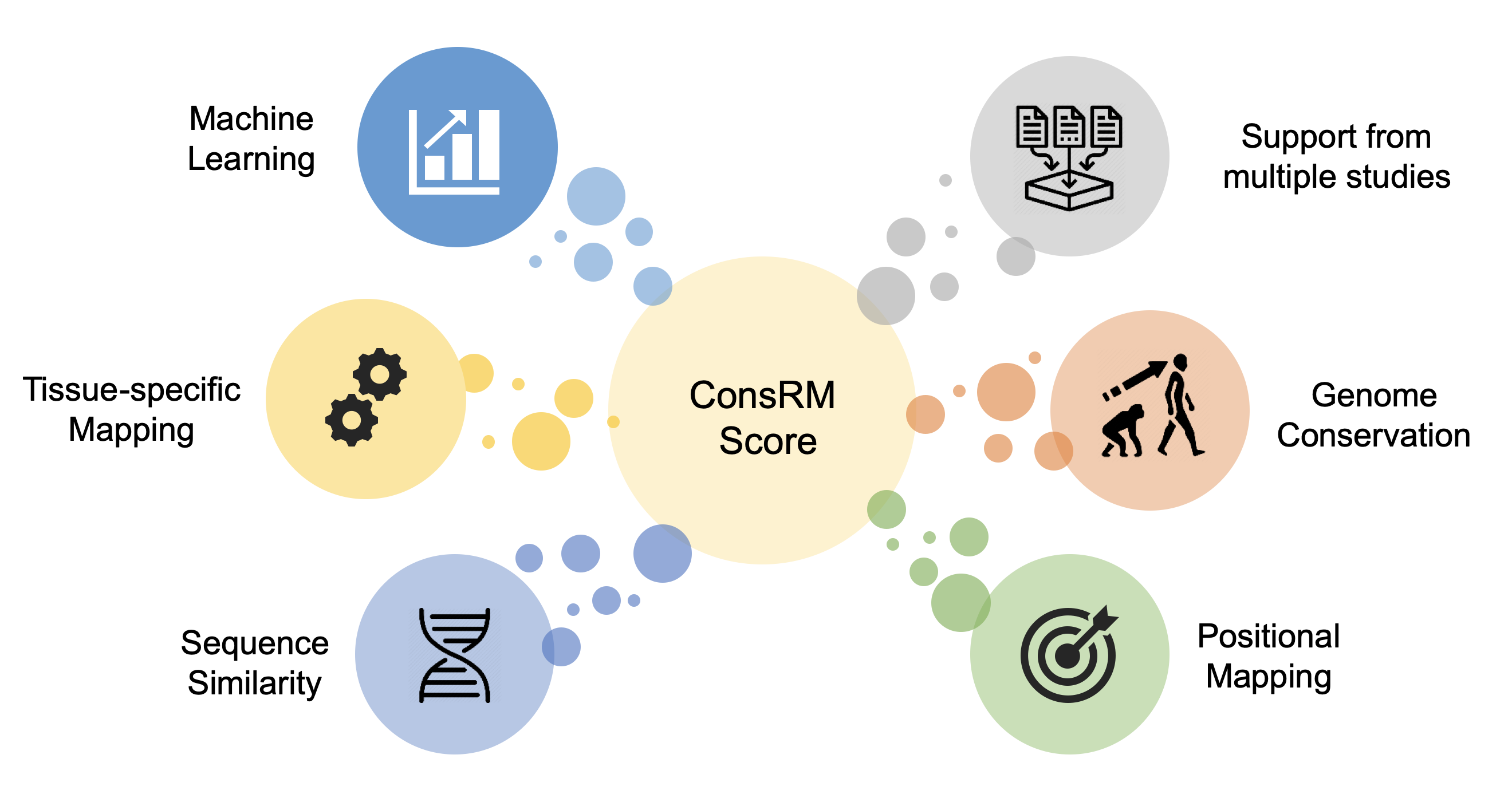
- N6-methyladenosine (m6A) is the most prevalent and abundant RNA modification on mRNAs and lncRNAs. Increasing evidences have revealed its crucial importance in essential molecular mechanisms and various diseases. With recent advances in sequencing techniques, tens of thousands of m6A sites are usually identified in a typical high-throughput experiment, posing a key challenge to distinguish the functional m6As from the rest of ‘passenger’ (or ‘silent’) ones. Given that functionally important m6As increase organismal fitness and are more likely to be conserved during evolution, we performed a comparative conservation analysis of the human and mouse m6A epitranscriptomes. Specifically, a novel scoring framework, ConsRM, was devised to quantitatively measure the degree of conservation of individual m6A RNA methylation sites, which including:
1): positional mapping
2): tissue-specific mapping
3): supports from multiple studies
4): sequence similarity
5): machine learning modeling
6): genome conservation

How to use database
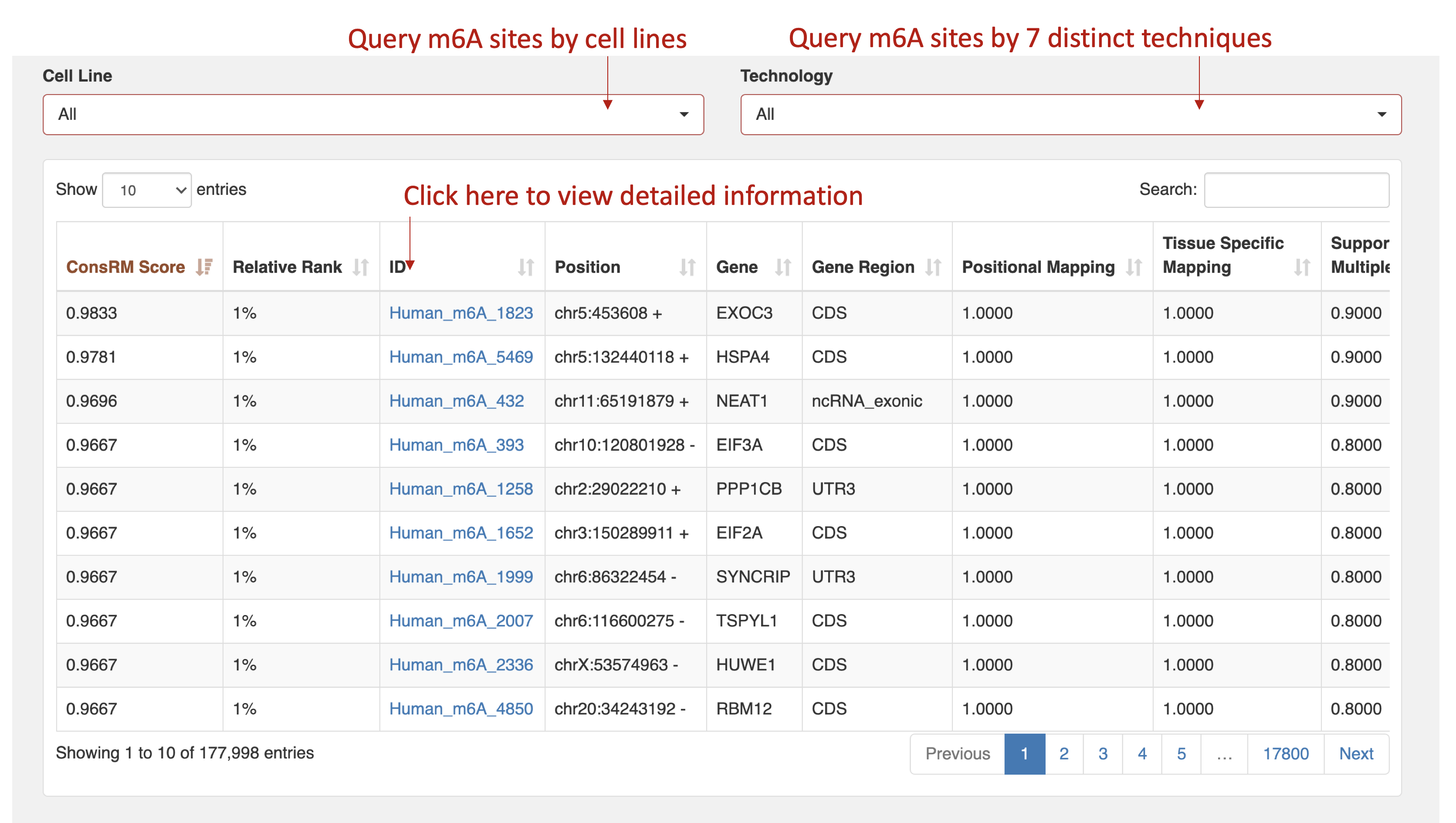
- To effectively share our findings, we developed a user-friendly online platform containing the conservation score of 177,998 human m6A RNA methylation sites, which conveys their potential epitranscriptome functionality. Users can query the database by different cell lines and high-throughput sequencing techniques.

Detailed information

- Specifically, the ConsRM score was calculated for each experimentally validated human m6A site by taking the average of the scores derived from the six aspects mentioned below, and ranges from 0 to 1.
- 1) Positional Mapping: Positional mapping concerns whether the RNAs transcribed from the conserved locus of human and mouse are simultaneously m6A modifiable at the corresponding residuals. For each human m6A site (based on hg19 genome assembly), its corresponding coordinate in mouse transcriptome (based on mm10 genome assembly) was identified using UCSC LiftOver tool. The conservation scores of human m6As were assigned for 1 mark if their corresponding coordinates in mouse transcriptome were also m6A modifiable. Besides precise positional mapping, we also checked whether the nearby regions for possible imprecise mapping, and assigned 0.8, 0.6, 0.4, and 0.2 mark for human m6As if an m6A modification were detected at 1bp, 2bp, 3bp and 4bp distance from their corresponding mouse coordinates.
- 2) Tissue-specific Mapping: Tissue-specific mapping concerns whether the RNAs transcribed from the conserved locus of human and mouse are simultaneously m6A modified at the same tissue. Currently, m6As were successfully identified under 12 tissues from human transcriptome, and for mouse, this number slightly dropped to 9. Among them, 4 tissues were shared by both species, including brain, liver, kidney and embryonic stem cell (ESC).
- 3) Supports from multiple studies: Supports from multiple studies concerns whether an m6A site can be detected by multiple m6A profiling studies. We suspected the m6As detected under multiple studies are more reliable with less possibility of being a false positive signal. For this reason, we assigned 1, 0.8, 0.6, 0.4, or 0.2 mark for m6As can be detected by more than 6, 5-6, 3-4, 2, or 1 m6A profiling datasets.
- 4) Sequence Similarity: Sequence similarity of m6A surrounding bases was also considered. Specifically, sequences were exacted from the 11bp flanking windows centered on a human m6A and its corresponding residual in the mouse transcriptome, respectively.
- 5) Machine Learning Model: Machine learning model was applied for inferring the conserved m6A sites.
- 6) Genome Conservation: Genome conservation. The phastCons 100-way conservation scores was calculated for human genome derived from genome-wide multiple alignments with other 99 vertebrate species, which was integrated into our scoring framework to evaluate the conservation degree of each human m6A site from a more general perspective.

How to use ConsRM web-server
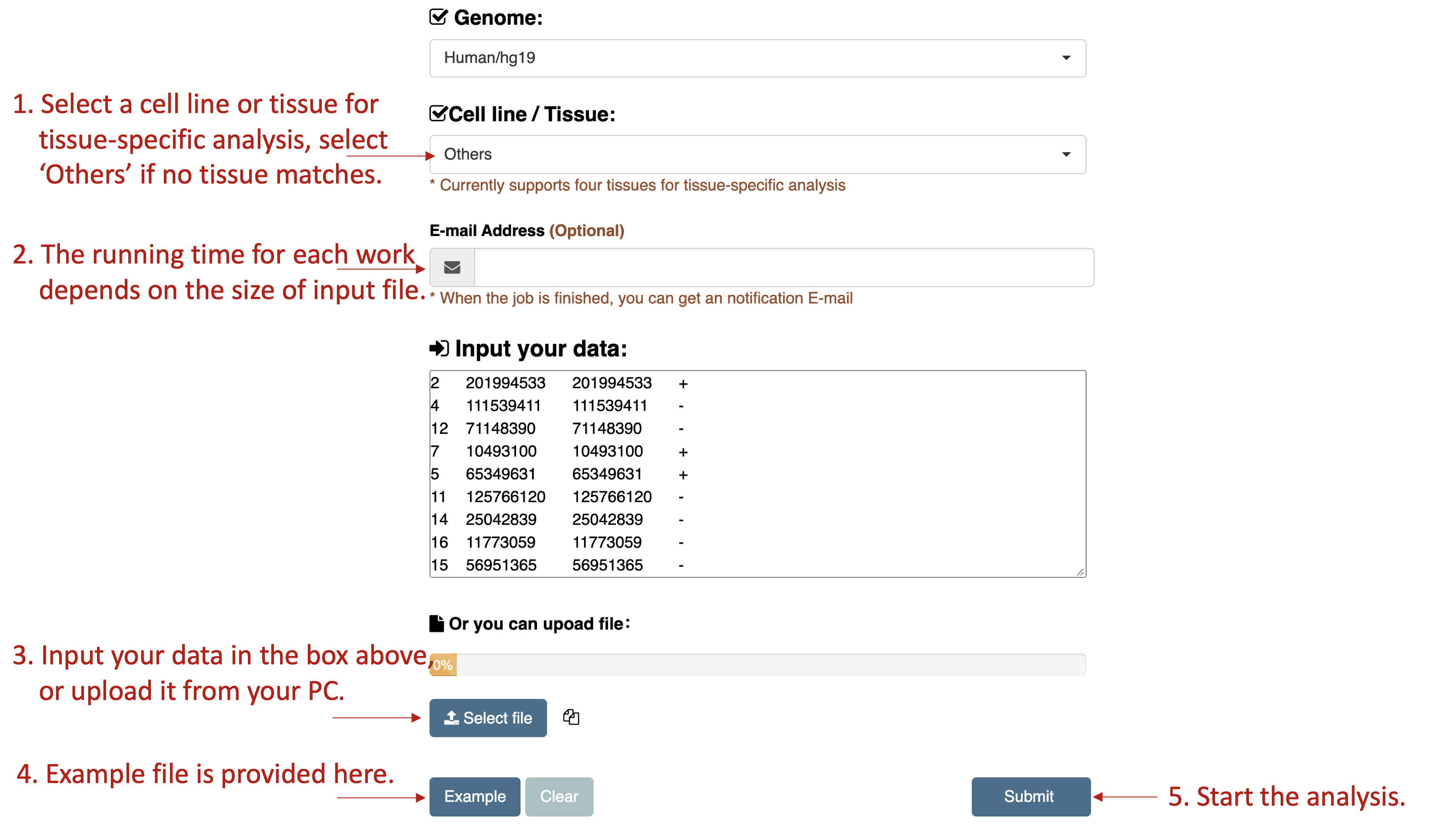
- A web server was constructed for calculating the ConsRM scores for a user provided list of m6A RNA methylation sites, with the statistical significance of the conservation score being assessed by comparing to all the experimentally validated m6As collected in the ConsRM database via the upper bound of the p-value.
