Introduction
m6 🅐 -Atlas
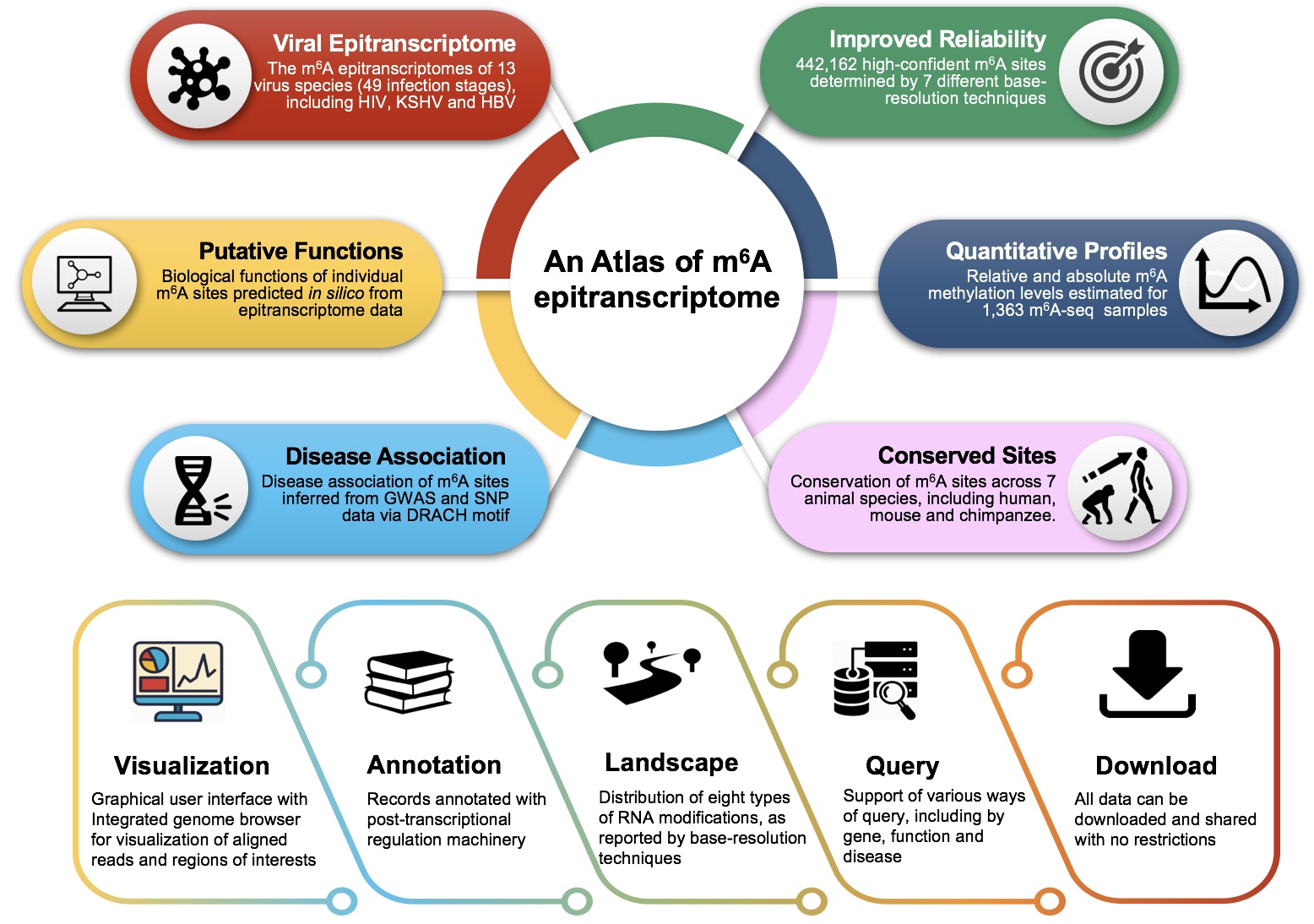
N6-methyladenosine (m6A) is currently the most prevalent RNA modification on mRNAs and lncRNAs, and plays a pivotal role during various biological processes and disease pathogenesis. Here, we developed m6A-Atlas, a comprehensive knowledgebase for unraveling the m6A epitranscriptome. (1) 442162 high-confidence m6A sites identified from seven base-resolution technologies were collected, and the quantitative (rather than binary) epitranscriptome profiles were estimated from 1363 high-throughput sequencing samples. (2) The conservation of m6A sites among seven vertebrate species, including human, mouse and chimp, and the 10 viruses’ m6A epitranscriptomes, including HIV, KSHV and DENV. (3) The putative biological functions of individual m6A sites predicted from epitranscriptome data, and the potential pathogenesis of m6A sites inferred from disease-associated genetic mutations that can directly destroy m6A directing sequence motifs. (4) Annotation of m6A epitranscriptome sites with post-transcriptional machinery (RBP-binding, microRNA interaction and splicing sites), and the landscape of multiple RNA modifications, together provided users to browse and do further studies.
Contents of m6A-Atlas
| Models |
 Human |
 Mouse |
 Rat |
 Zebrafish |
 Fly |

Arabido-psis
|
 Yeast |
|---|---|---|---|---|---|---|---|
| Reliable m6A Sites | |||||||
| Quantitative Profiles | |||||||
| Basic Annotation | |||||||
| Conservation in Vertebrates | |||||||
| Putative Functions | |||||||
| Landscape of Modifications | |||||||
| Disease Association |
User guide
Getting Started
We present here m6🅐-Atlas, a centralized online platform for a high-confidence collection of unique m6A sites. The m6A-Atlas consists of the following two major components, including: eukaryote and virus m6 A site databases. On the Home Page, the Quick search feature allows users to select seven species and find m6A sites using a search query (i.e. Gene, Region or Disease-Associationenomic location). Quick search is available by using the search bar; The

Eukaryote
1. Eukaryote Table
m6A-Atlas provides users a webpage for a high-confidence collection of reliable m6A sites identified from seven base-resolution technologies covering seven eukaryotes. The detail list of m6A sites, corresponding cell lines and tissues, relative technologies and the statistical results were summarized and presented, according to the needed filter selection of species, RNA modification, cell line, and technique (e.g. Human).

2.Basic Info
Users can click “Site ID” to see more information. On Basic Info section, users click on button, to link to Jbrowser showing m6A-sites, gene, RNA Binding region, miRNA target and other RNA modfication sites.
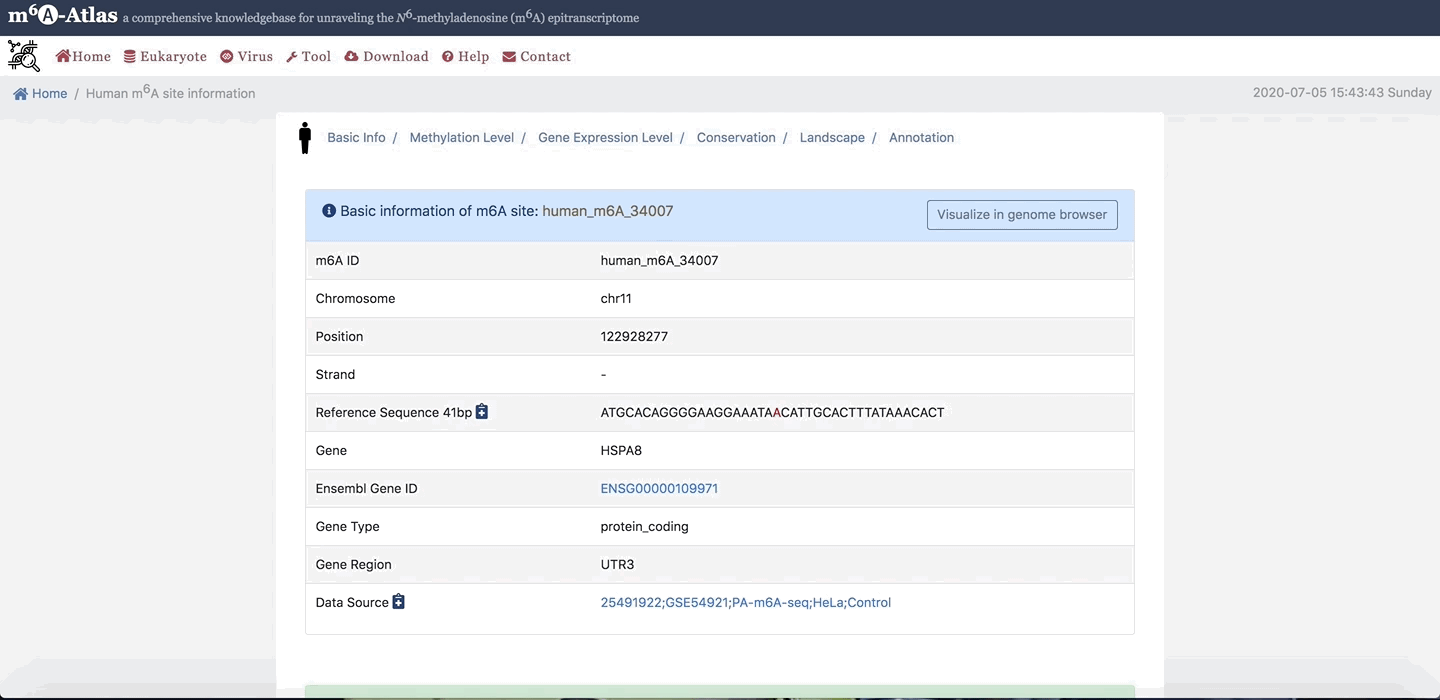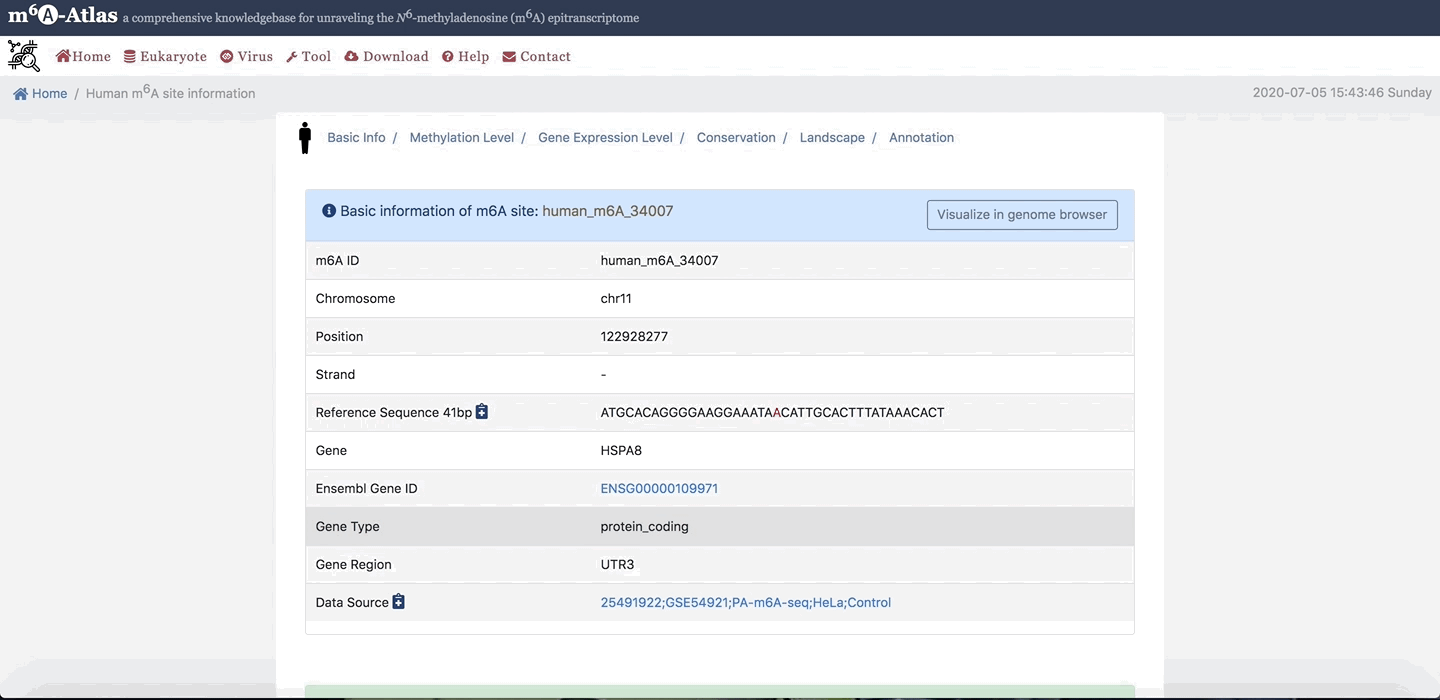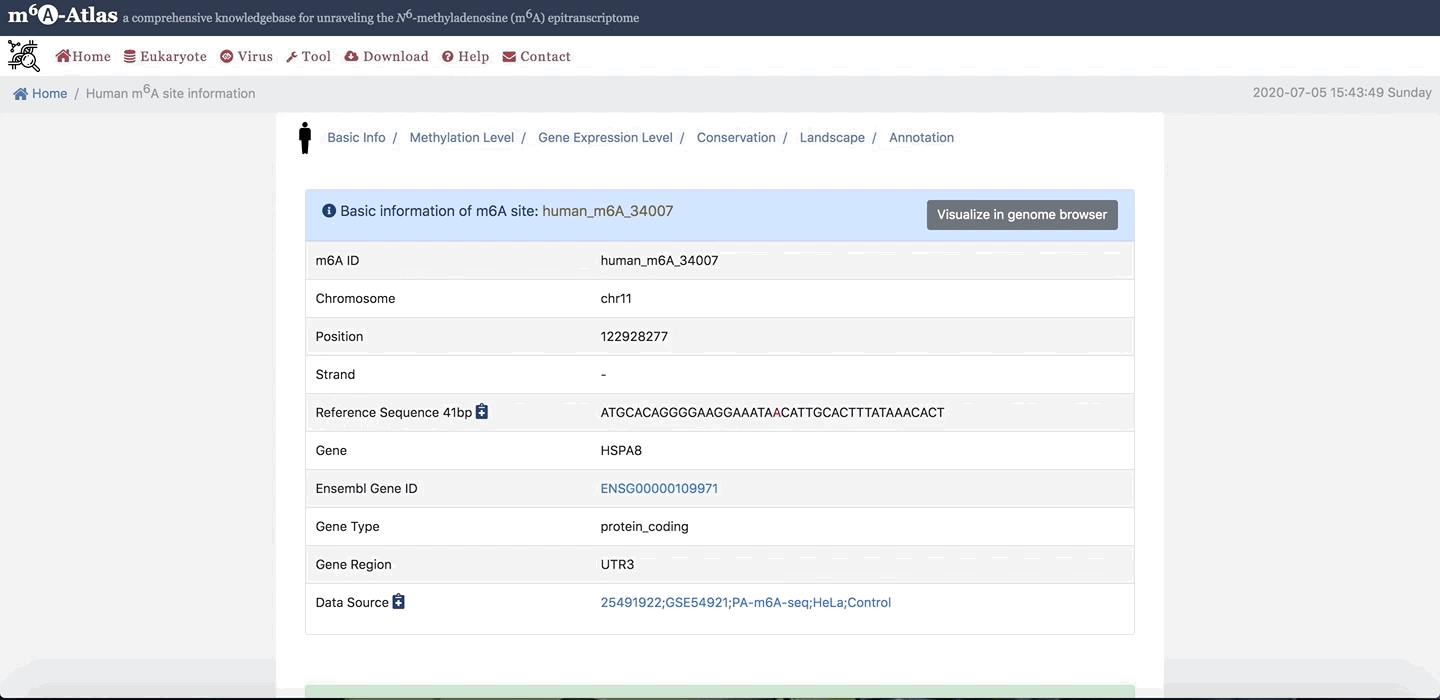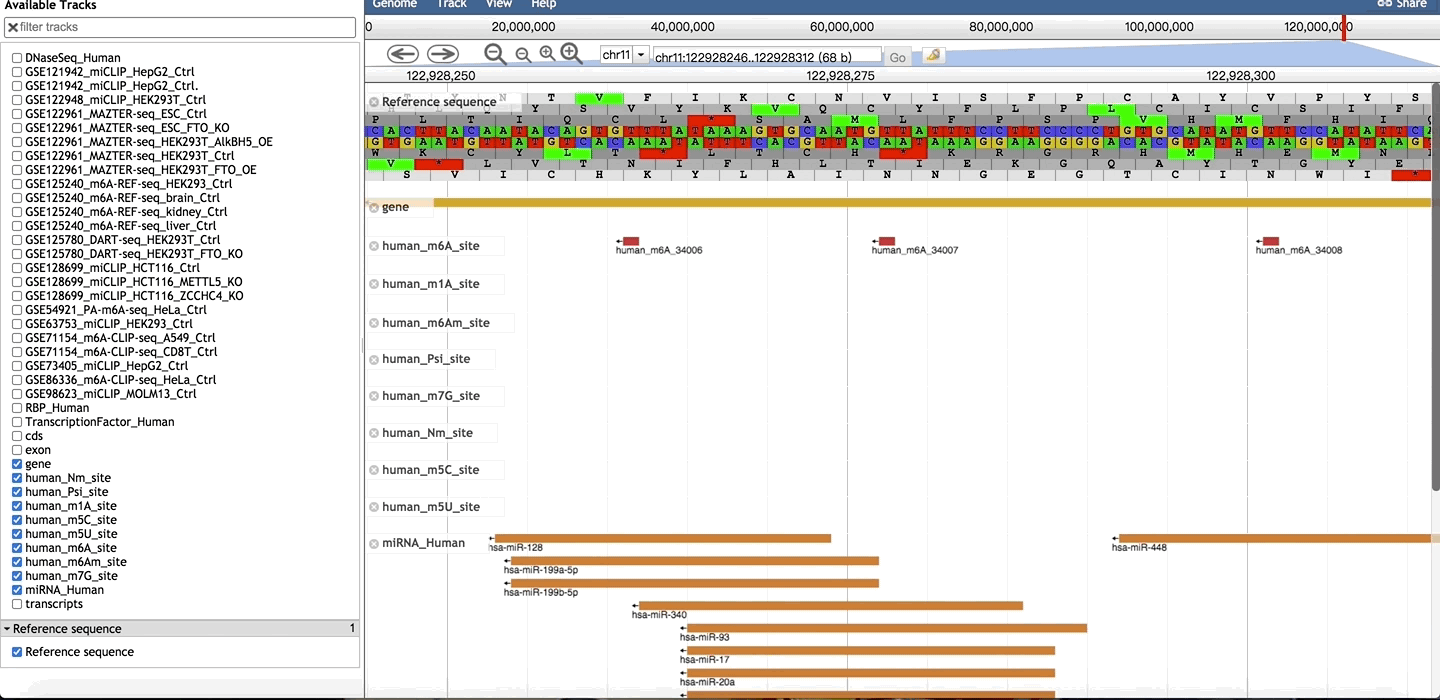
3. Methylation Level
m6A-Atlas provides users an interactive drawing to show the differential methylation levels under various conditions. The first choice is to select the methylation level on normal cell line; the second choice is to display the methylation level after virus infection. Users can select functions of several choices to query the various cell lines, techniques or viruses which you are interested in.

4. Gene Expression Level
Similarly, m6A-Atlas also provides users an interactive drawing to show the differential expression gene levels where m6A site(s) located under various conditions. The first choice is to select the gene expression level on normal cell line; the second choice is to display the gene expression level after virus infection. Users can select functions of several choices to query the various cell lines, techniques or viruses which you are interested in.

5. Conservation
Here, m6A-Atlas also offers conservation analysis was performed among human, mouse, rat, zebrafish, pig, monkey and chimpanzee to identify the conserved m6A sites between two vertebrate species.
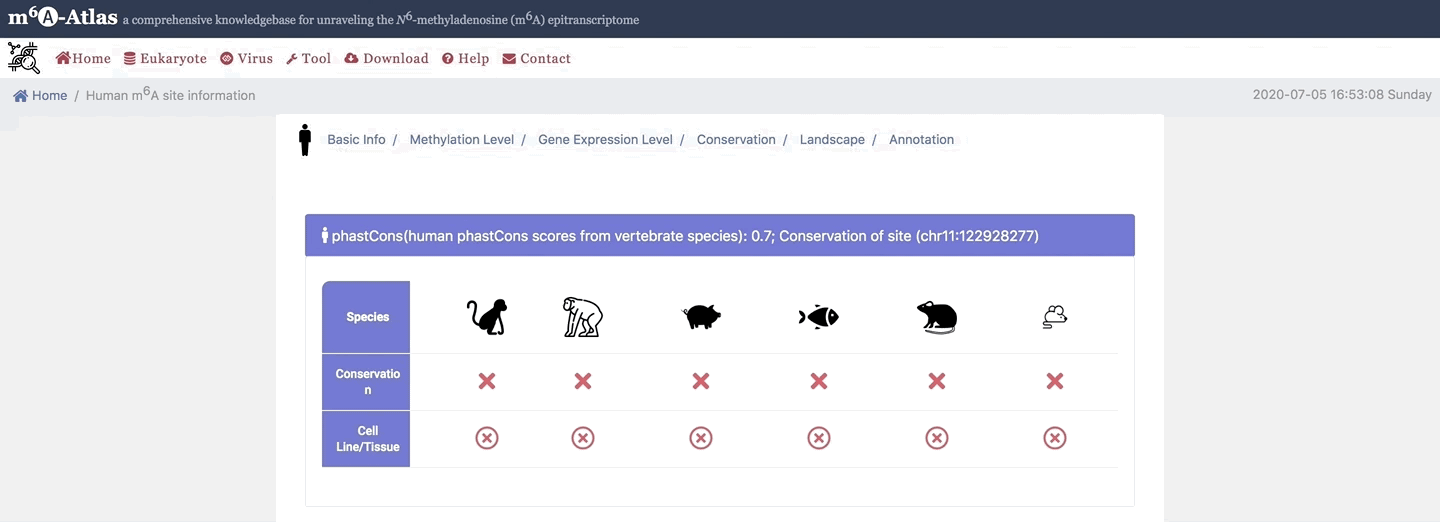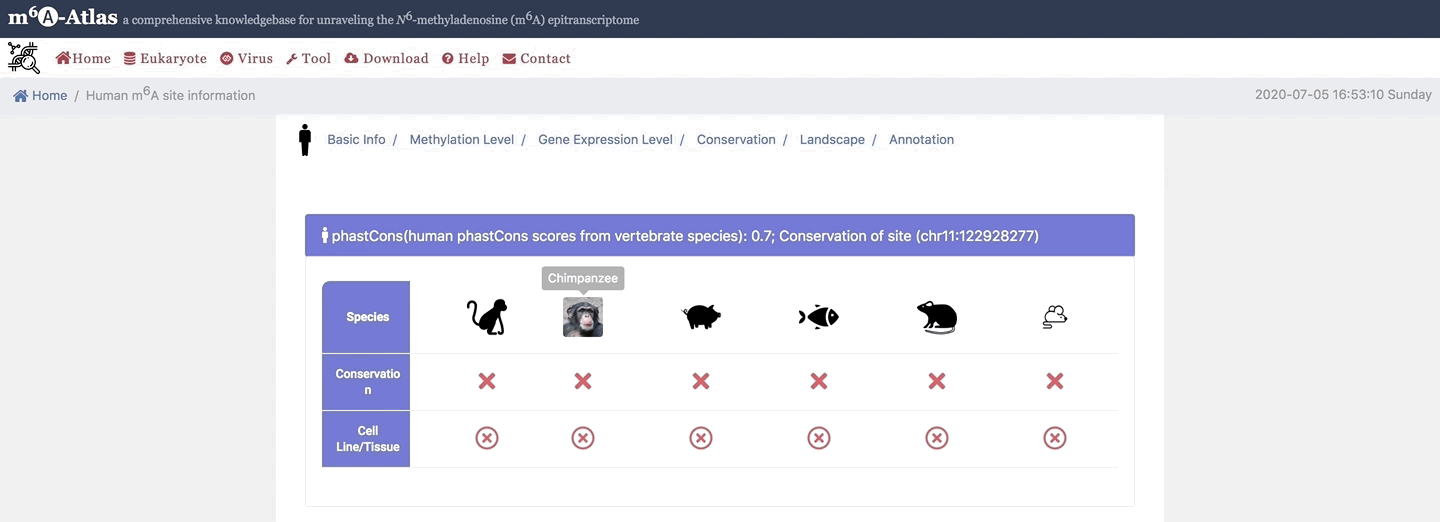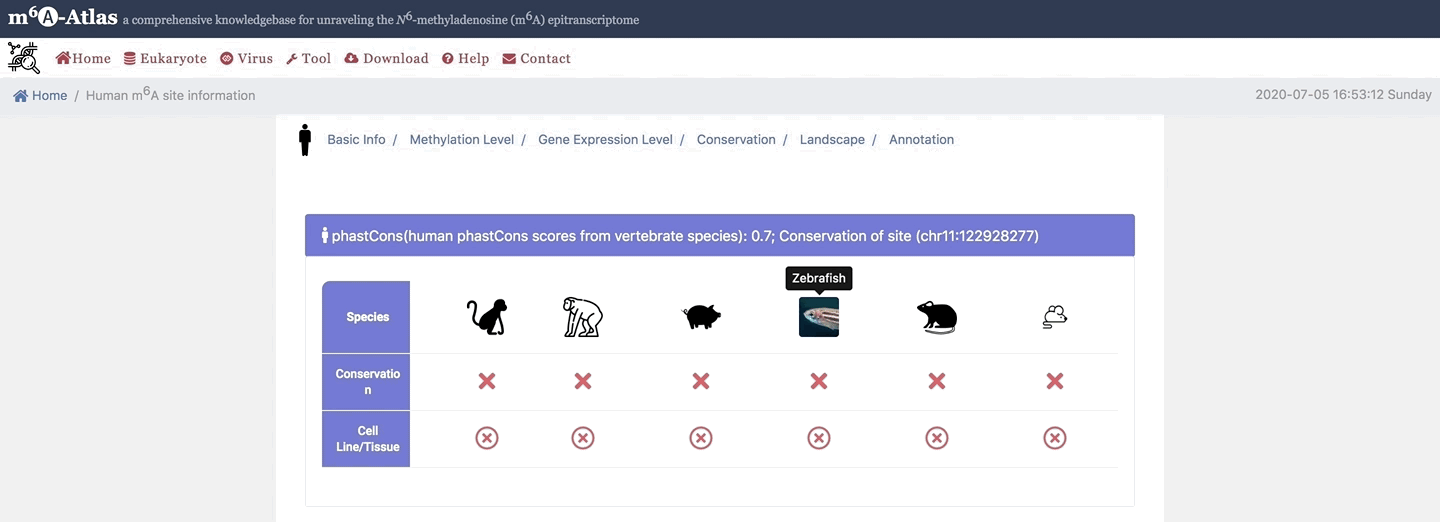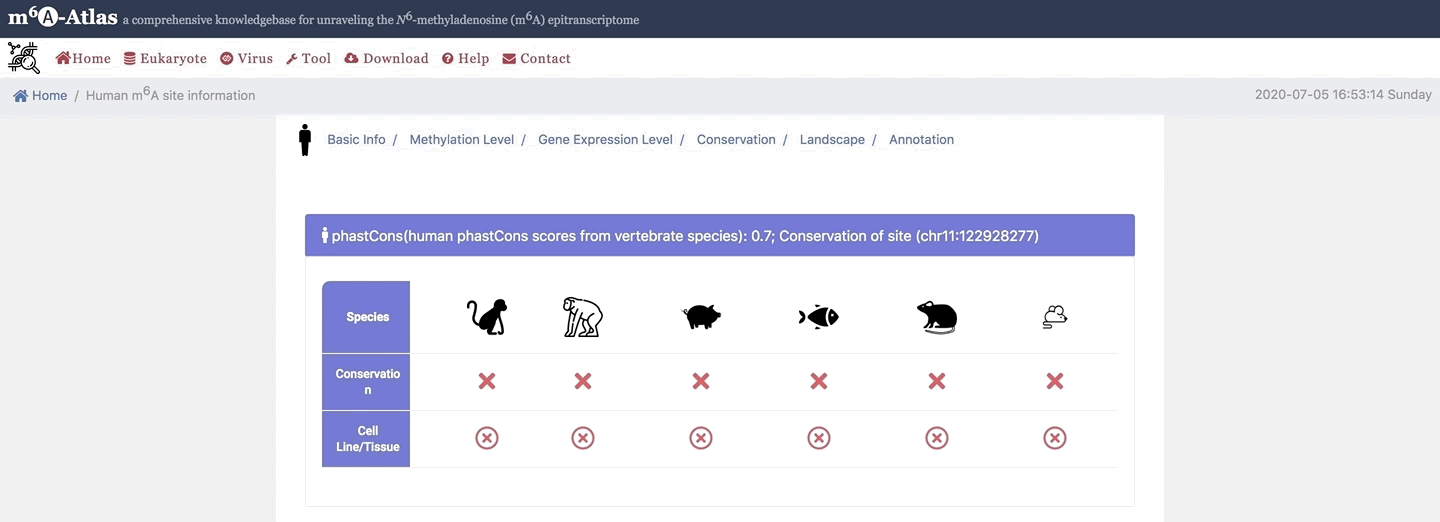
6. Landscape
m6A-Atlas provides users an interactive RNA modfication landscape, where gene regions and different RNA modification sites distribution plots are demonstrated and it also show the annotation information involving in m6A sites shown at the top. Users can view the summary table for the number of m6A sites and their closer RNA modification sites by clicking the Table button and downloading this table.

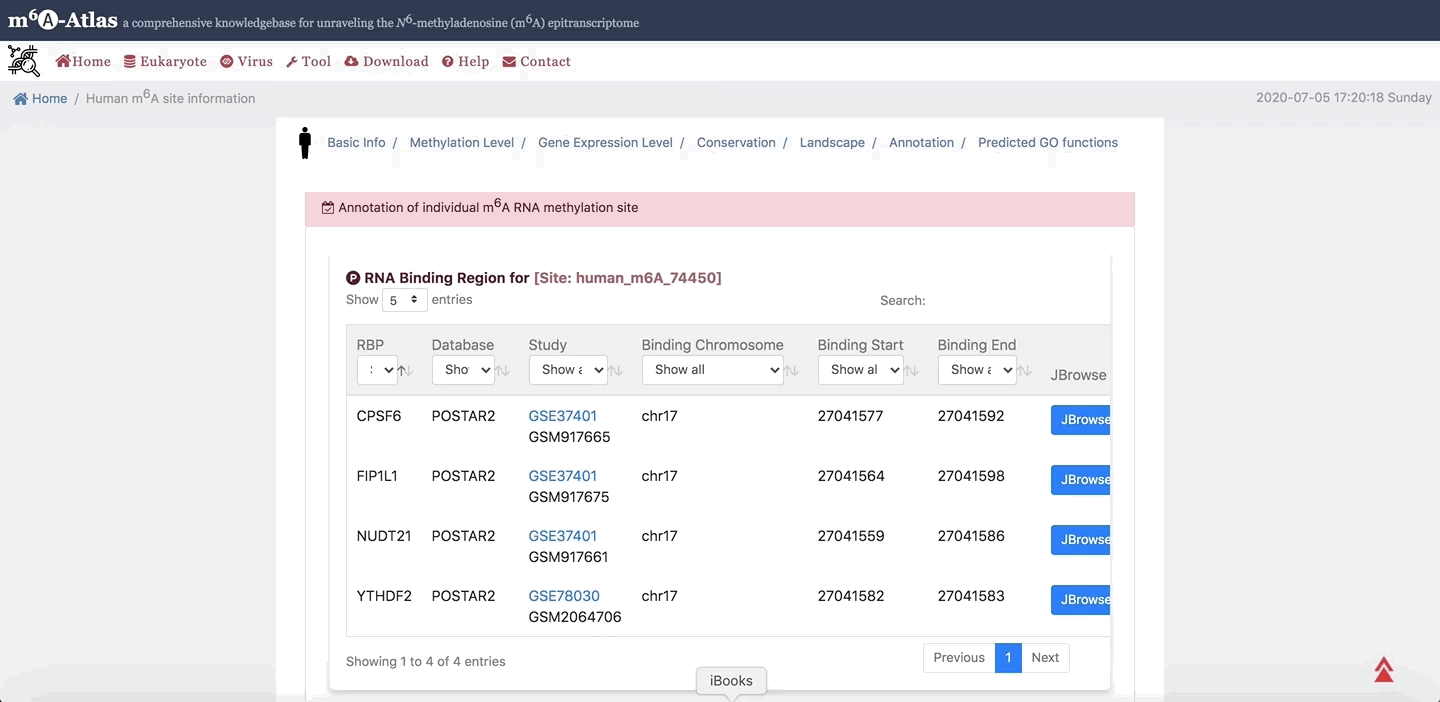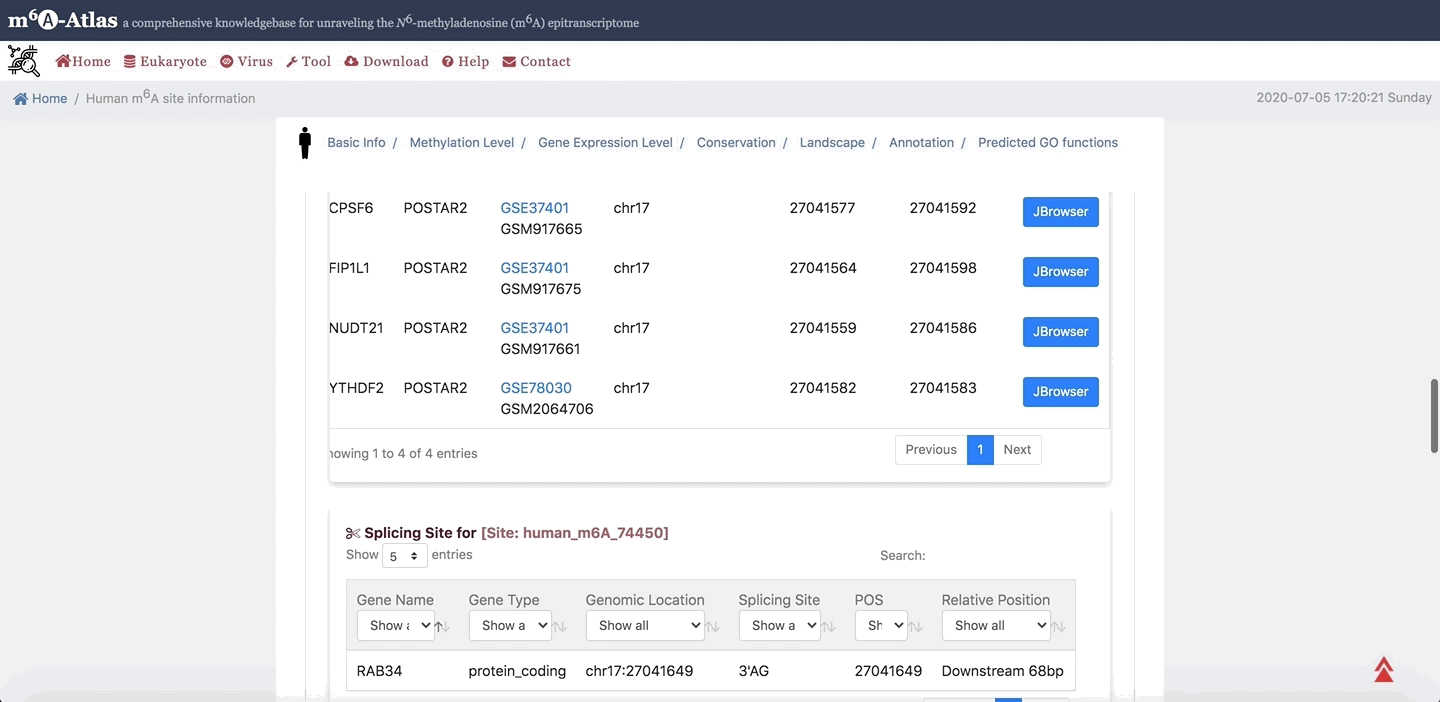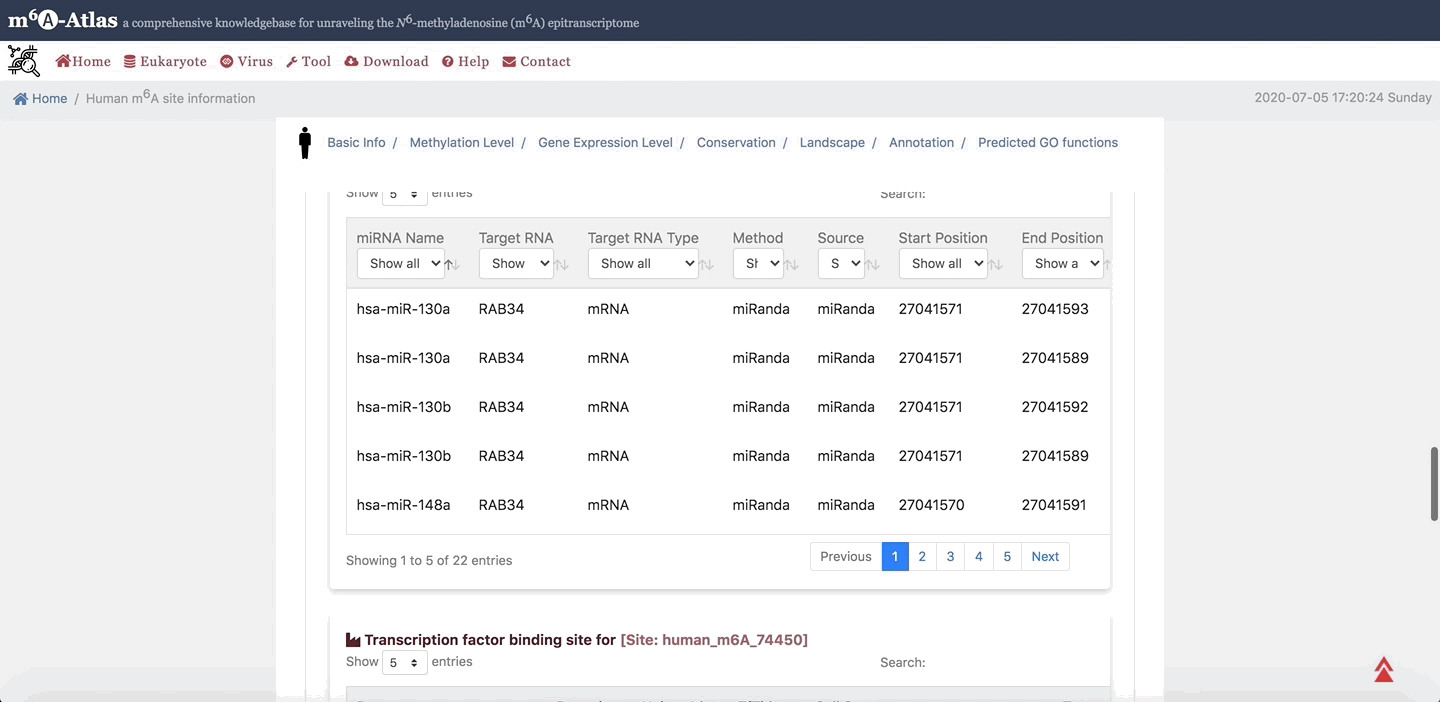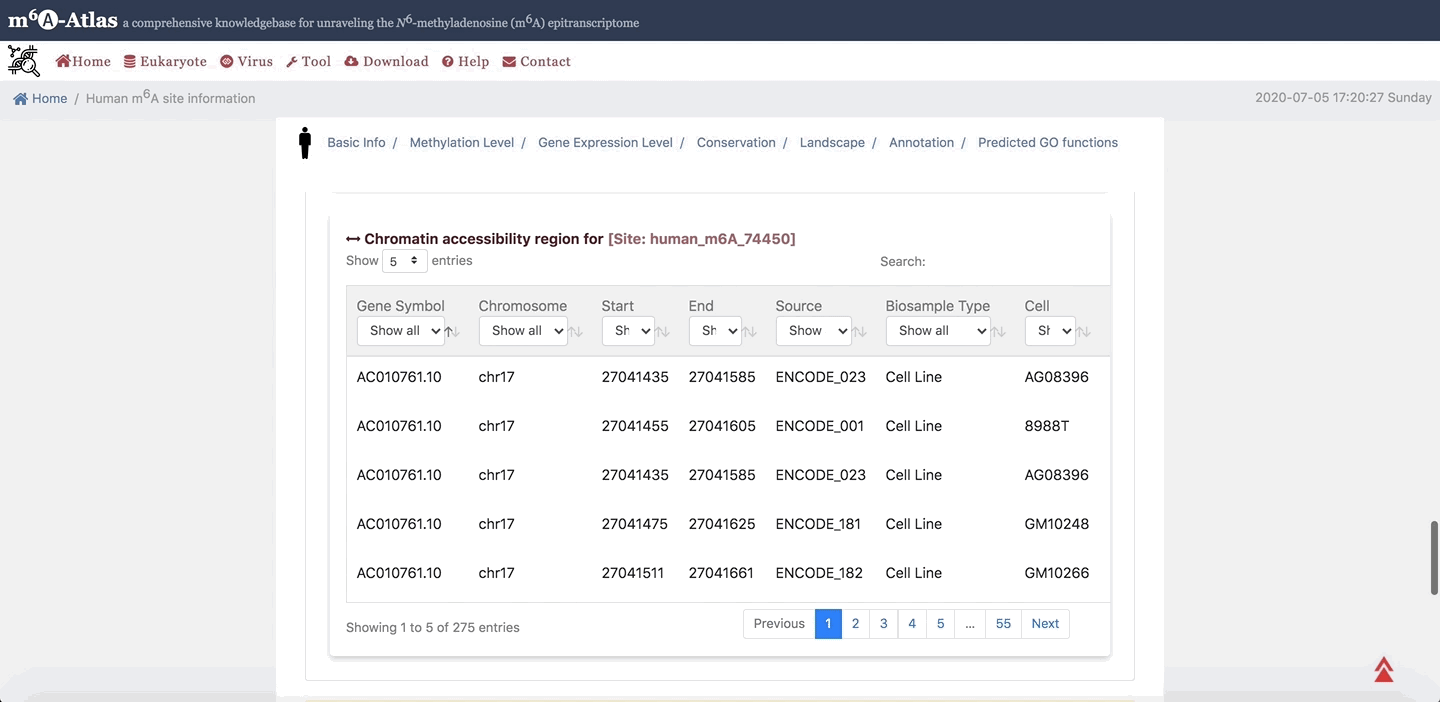
7. Annotation
m6A-Atlas provides the basic annotation for m6A sites, including the splicing sites, miRNA target sites, RBP-binding sites and subcellular location integrated to help understand the regulatory roles of m6A. Moreover, the potential pathogenesis of human m6A sites inferred from disease-associated genetic mutations that can destroy m6A forming sequence motif are also demonstrated.
8. GO prediction
m6A-Atlas makes the putative biological functions of individual m6A site were predicted for the conserved between human and mouse according to guilt-by-association principle, as previously described. On the GO predication section, the two tabular listings show neighbor sites of target m6A site and related predict GO functions. Click to see Network Graph of network ralation between the neighbor m6A site and GO functions.

Virus
1.Virus Table
Try to scroll this section and look at the navigation bar while scrolling!

Tool
1. m6A-conservation-finder
m6A-Atlas provides users a convenient Web Server to do a batch query. Among the m6A-conservation-finder is used to identify the conserved m6A sites among a user-provided list of candidate sites. According to inputted data or uploaded file (required bed format),and the corresponding samples are demonstrated; Users can choose different species (assemblies) including Human/hg19, Mouse/mm10, Rat/rn6 and Zebrafish/danRer10. It is noted that users are required to enter their E-mail address to receive a notification after their job is completed. After submitting , each job will be assigned a unique ID to the users. The job can be retrieved by waiting on the pop-up results webpage directly.

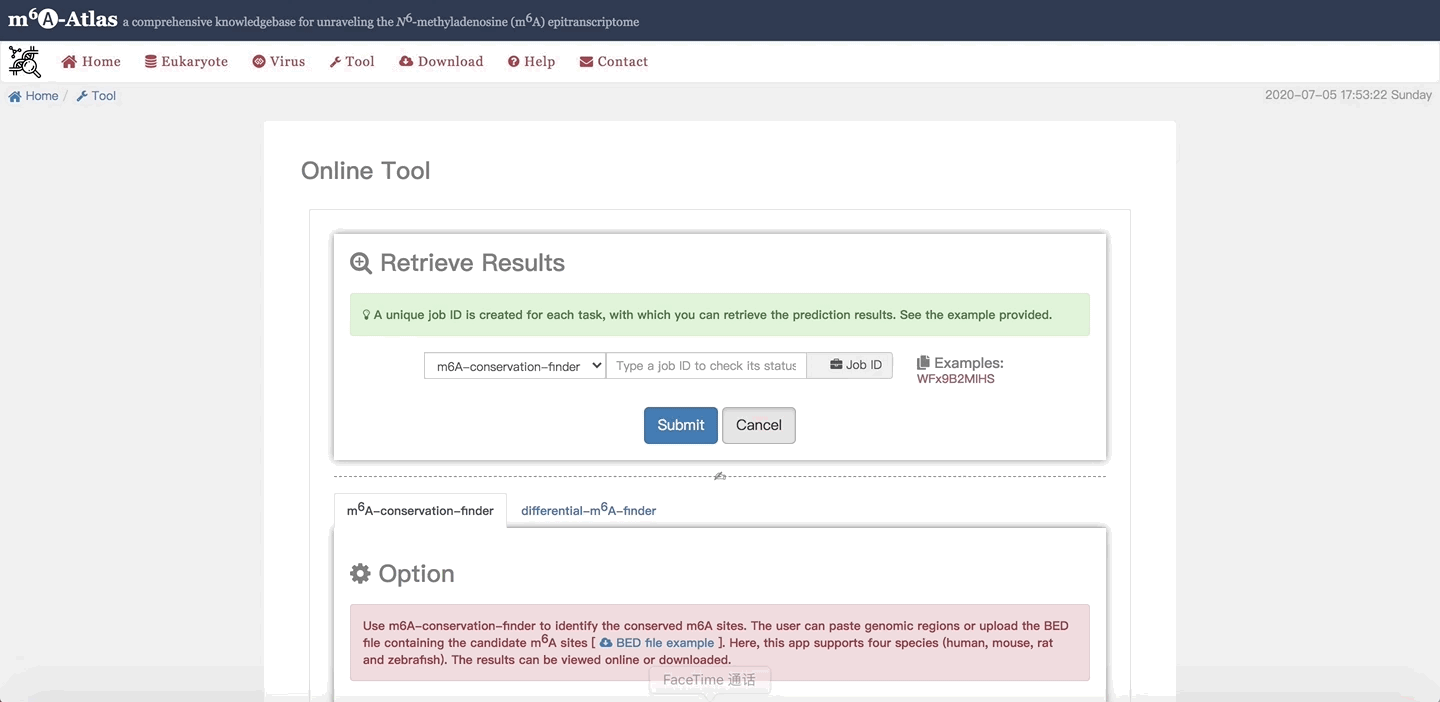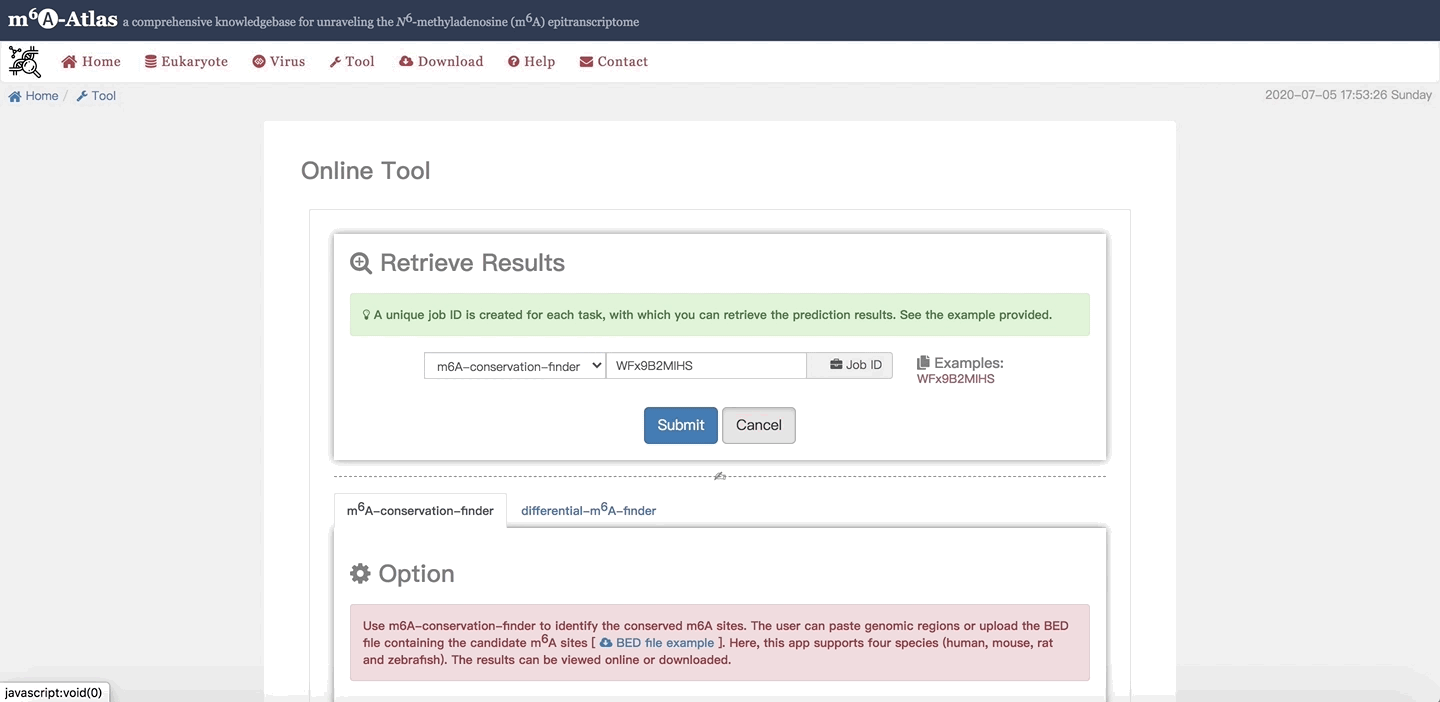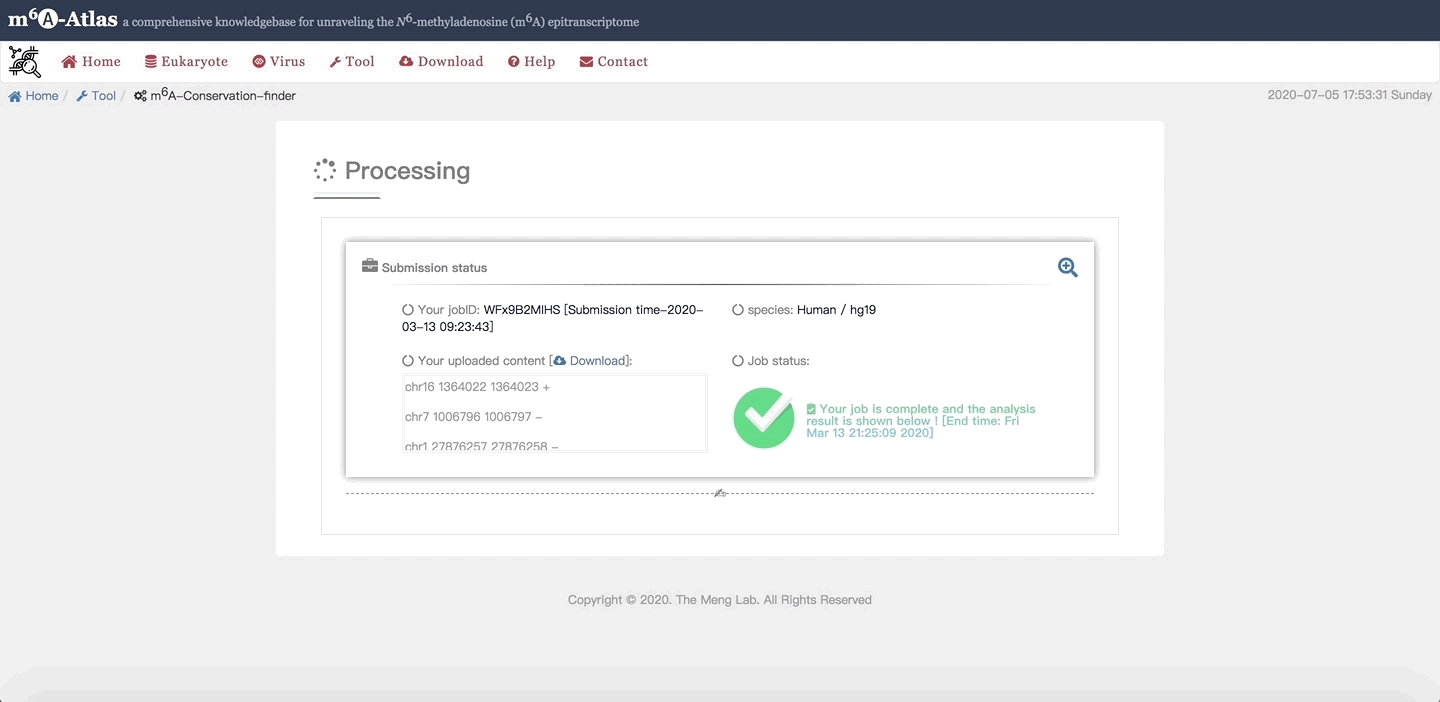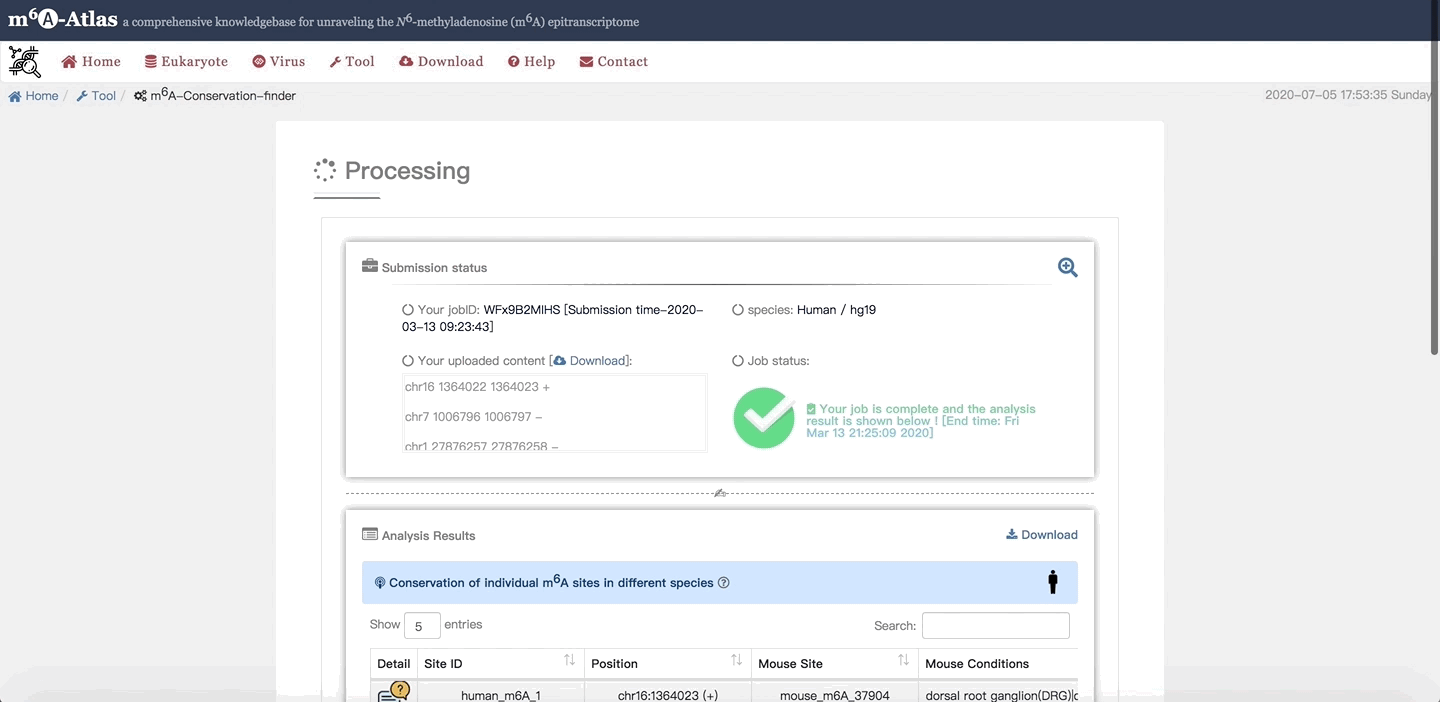
After the work is finished, the detail list of query species with predicted conservation sites in different species as well as related information are presented. Click on Download in the upper right corner, users can get their predicted results. Under the table, statistic analysis of predicted results is also illustrated.

2. differential-m6A-finder
m6A-Atlas provides users a convenient Web Server to do a batch query. Fot the differential-m6A-finder, it returns the differentially methylated m6A sites between two experimental conditions. Users can have several choices including the species and two different conditions, i.e. Homo sapiens (Human), GSE99017_brain_cortex_Ctrl and GSE128443_HeLa_Ctrl. Users can also look up the following table to select their interested conditions. It is noted that users are required to enter their E-mail address to receive a notification after their job is completed. After submitting, each job will be assigned a unique ID to the users. The job can be retrieved by waiting on the pop-up results webpage directly.

After the work is finished, the detail list of query specie with the differentially methylated m6A sites between two experimental conditions as well as related information are presented. Click on Download in the upper right corner, users can get their predicted result.

3. Retrieve Results
When users leave the job page or lose it by accident, the job ID can be used to retrieve their job.
Download
The data used on this website from m6A-Atlas online data and Quantification of m6A methylation levels is available and the statistic results were summarized and presented at bottom.