Introduction
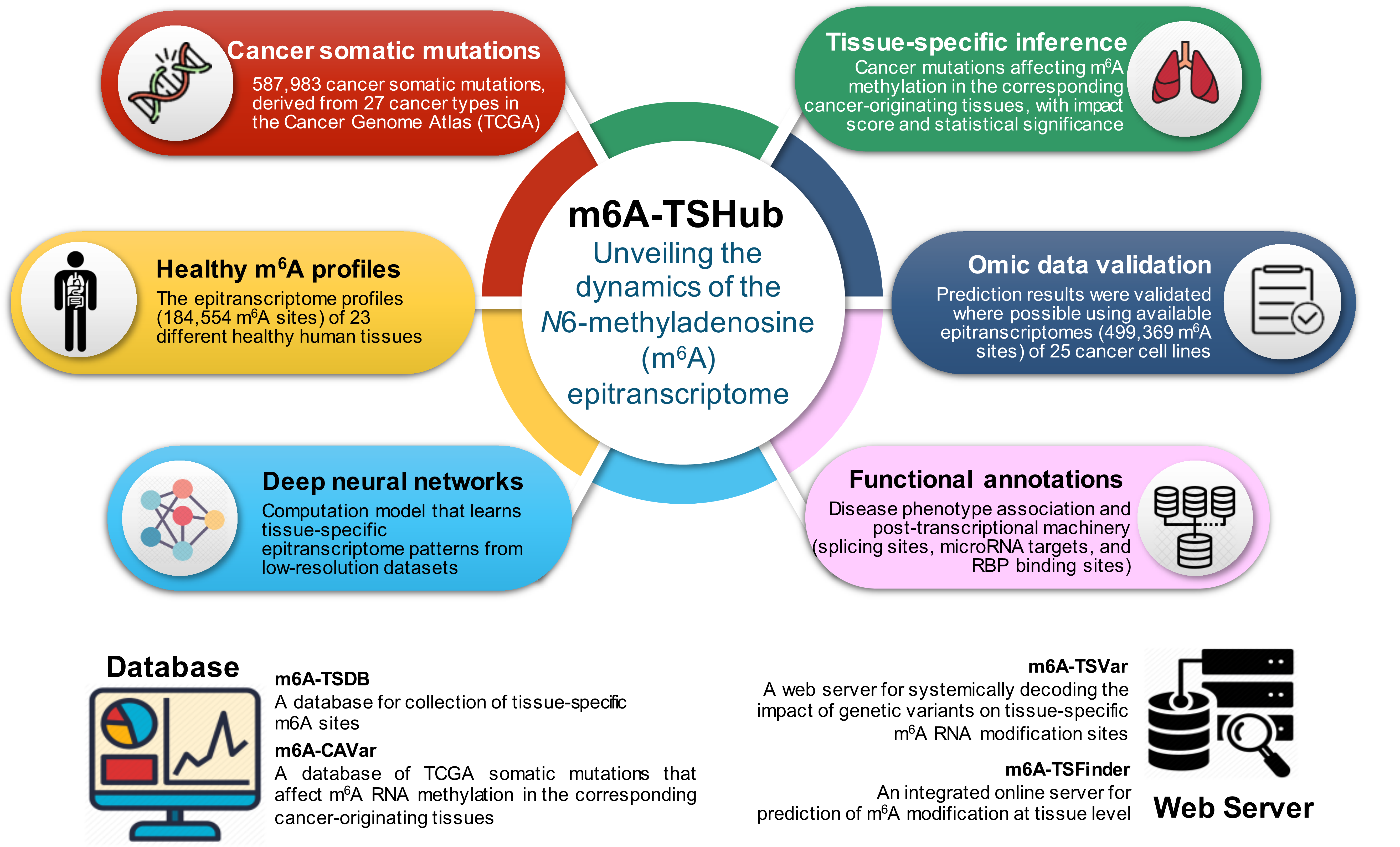
- As the most pervasive marker present on mRNA and lncRNA, N6-methyladenosine (m6A) RNA methylation has been shown to participate in various biological processes. Recent studies revealed the distinct patterns of m6A methylome across human tissues, and a major challenge remains in elucidating the tissue-specific presence and circuitry of m6A methylation. We present here a comprehensive online platform m6A-TSHub for unveiling the context-specific m6A methylation and m6A-affecting mutations in 23 human tissues. m6A-TSHub consists of four core components, including:
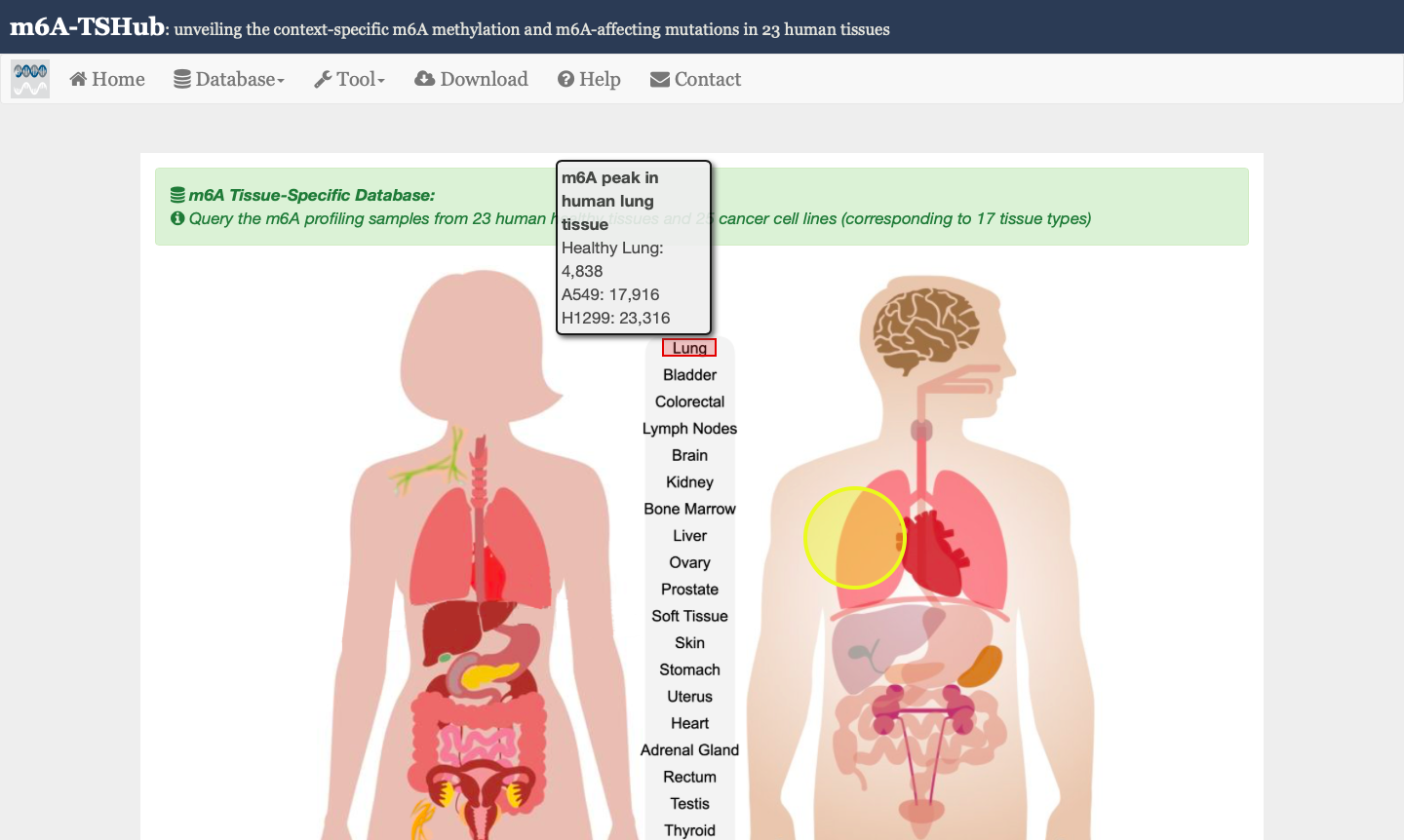
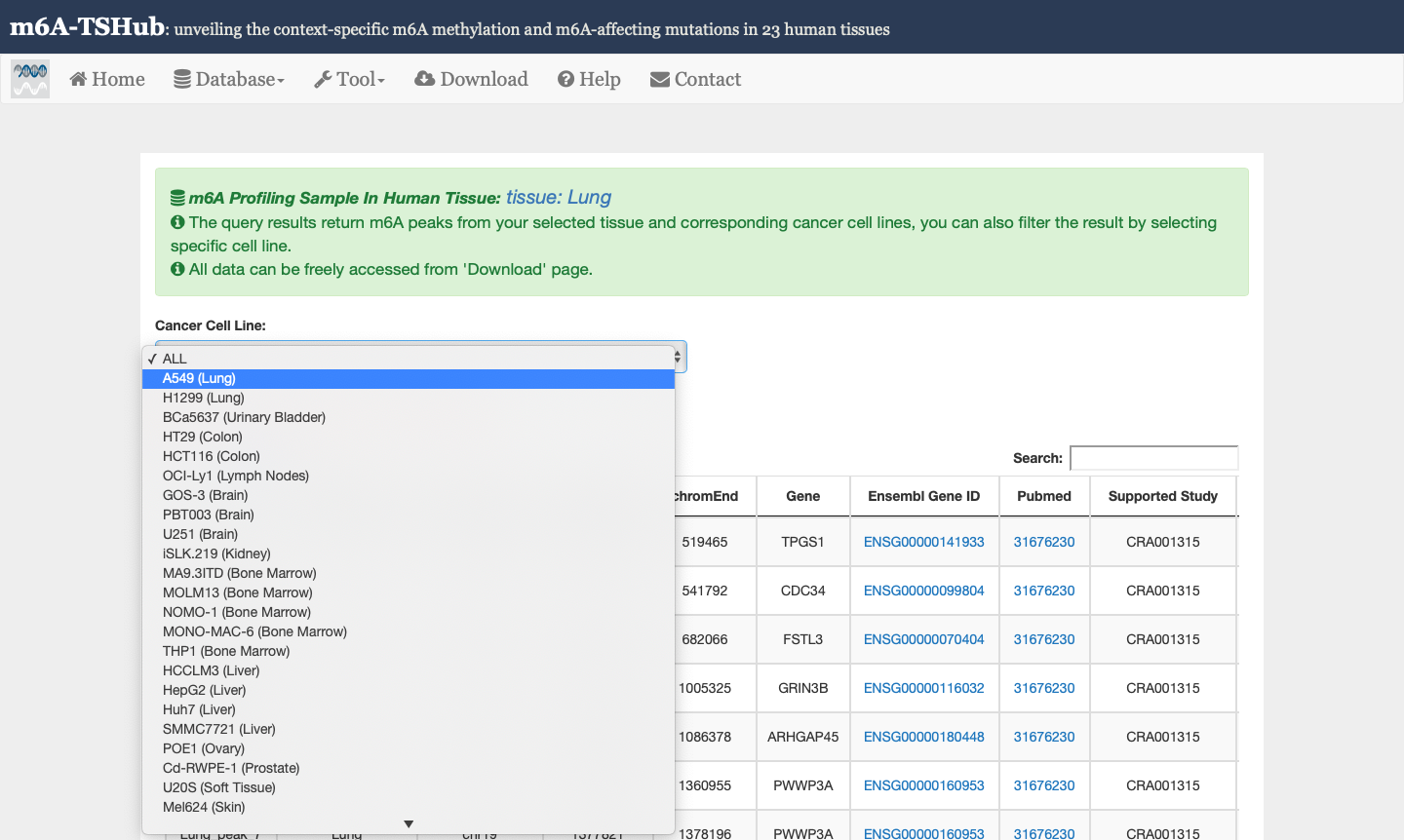
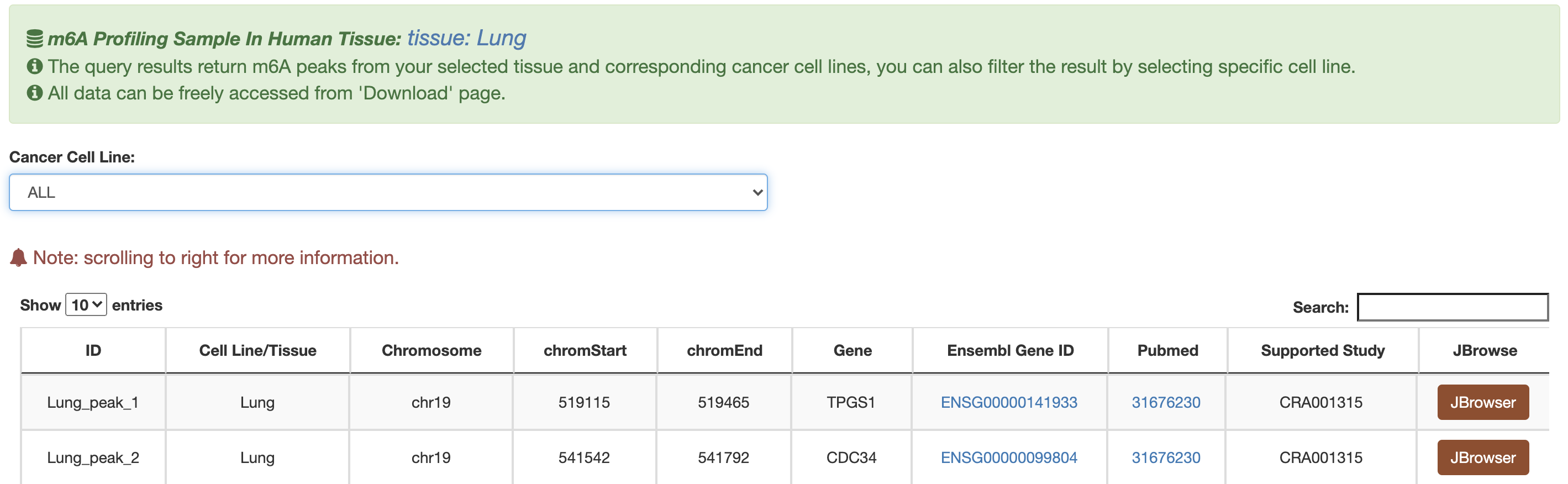
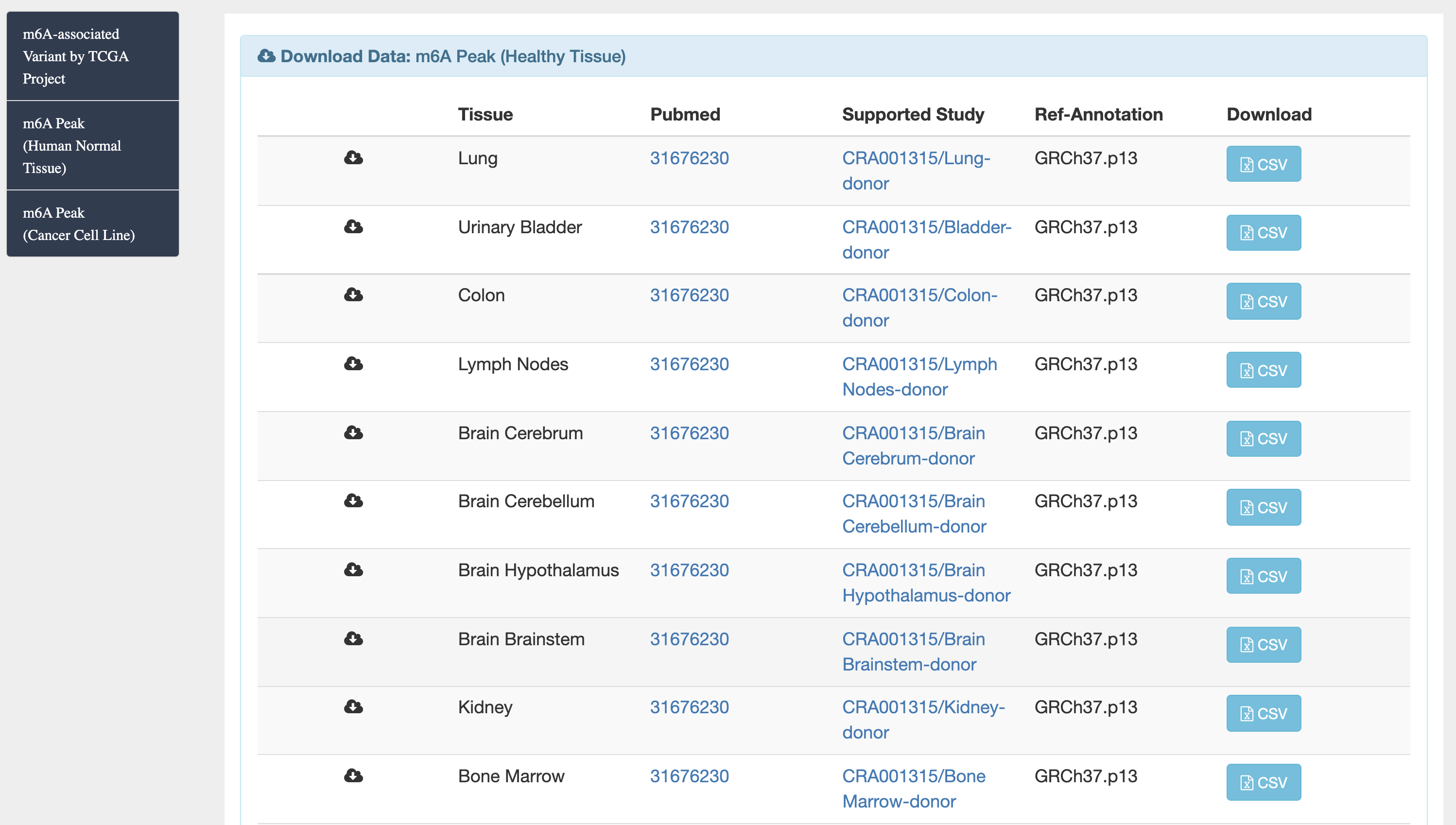
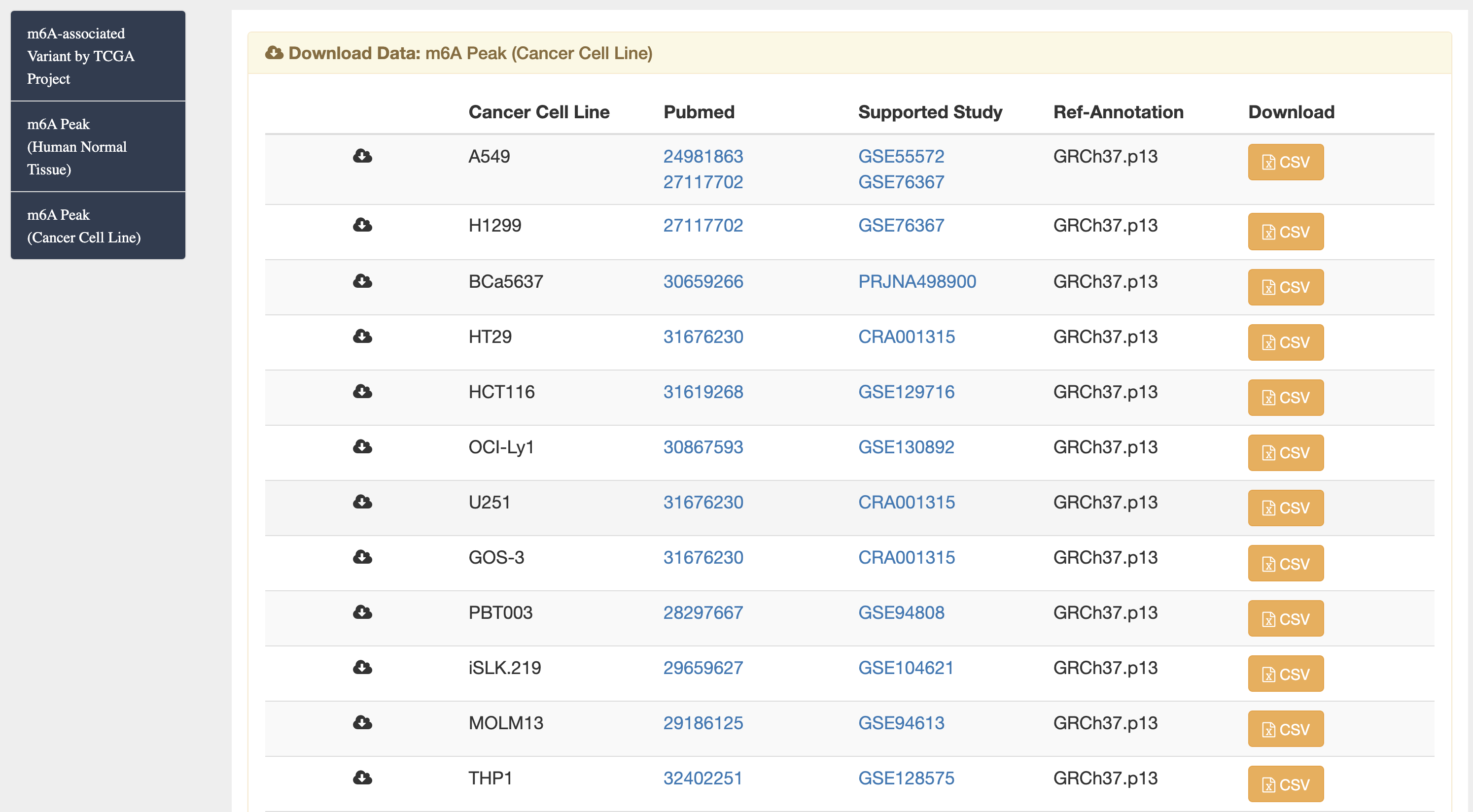
1) m6A-TSDB: a comprehensive database of 184,554 and 499,369 m6A-containing peaks collected from 23 normal human tissues and 25 cancer samples, respectively. Among them, 17 out of 25 tumor samples have the m6A profiles of their matched primary tissues.
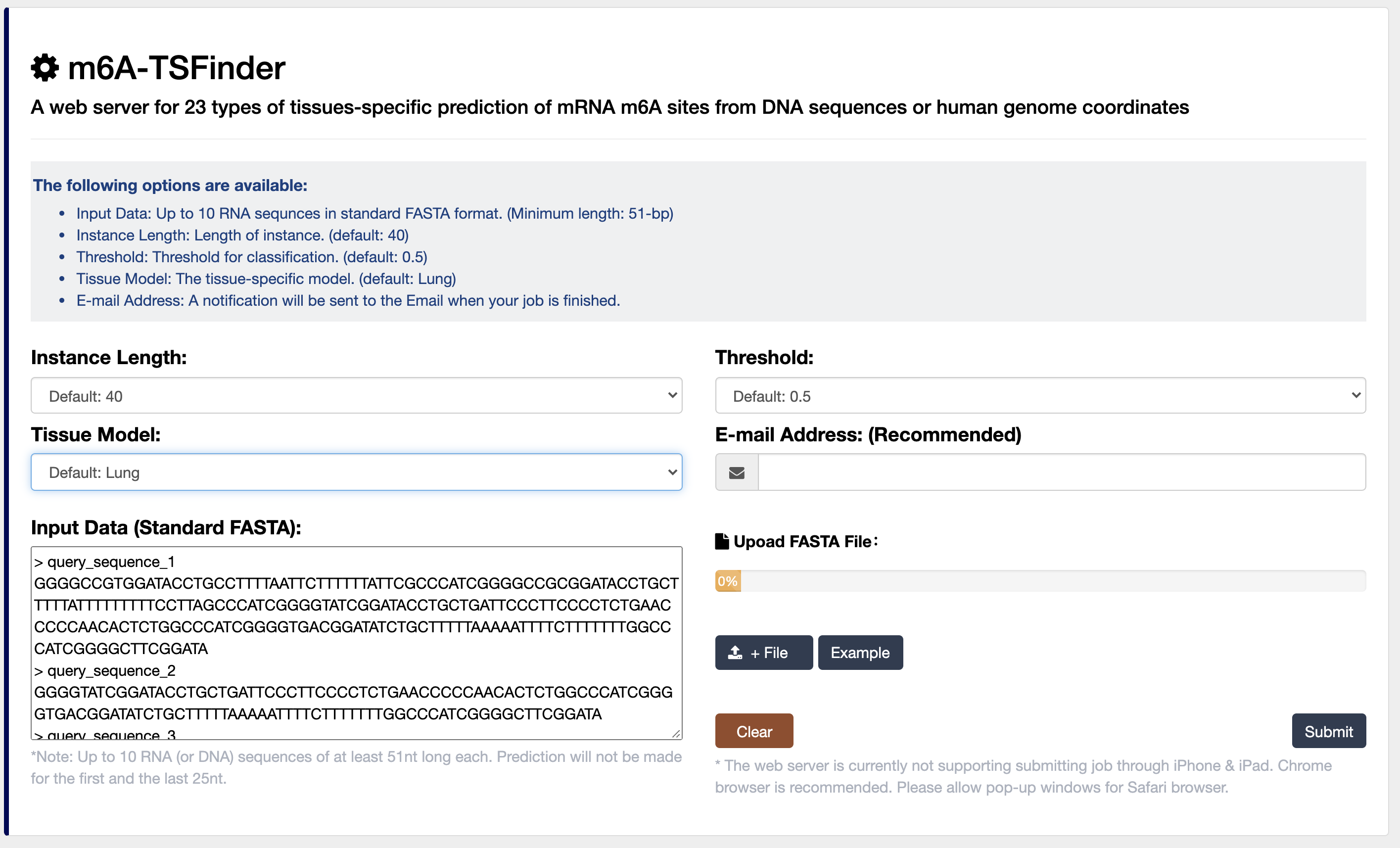
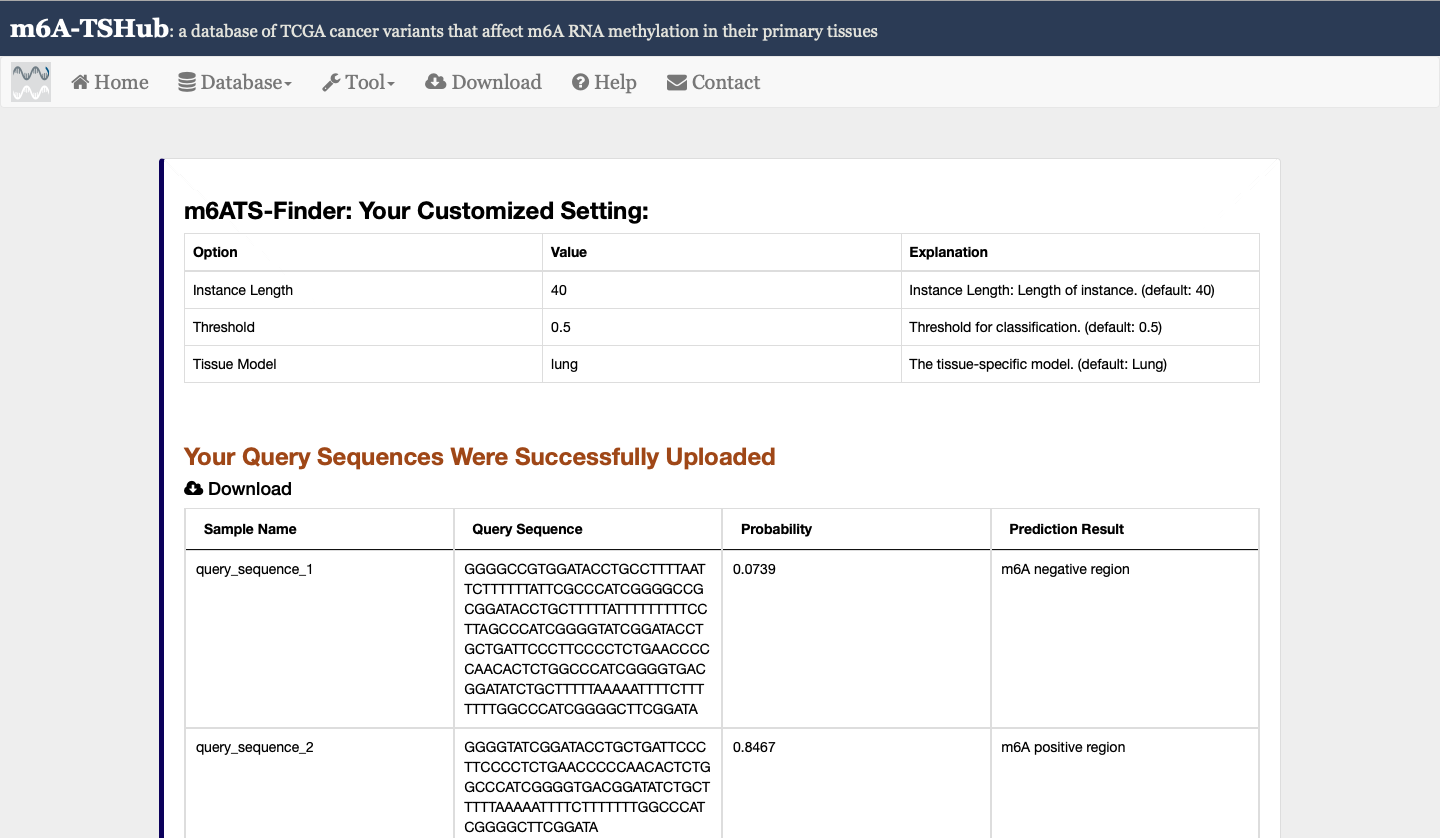
2) m6A-TSFinder: an integrated online server for the prediction of tissue-specific m6A modification in 23 human tissues, built upon a gated attention based multi-instance deep neural networks.
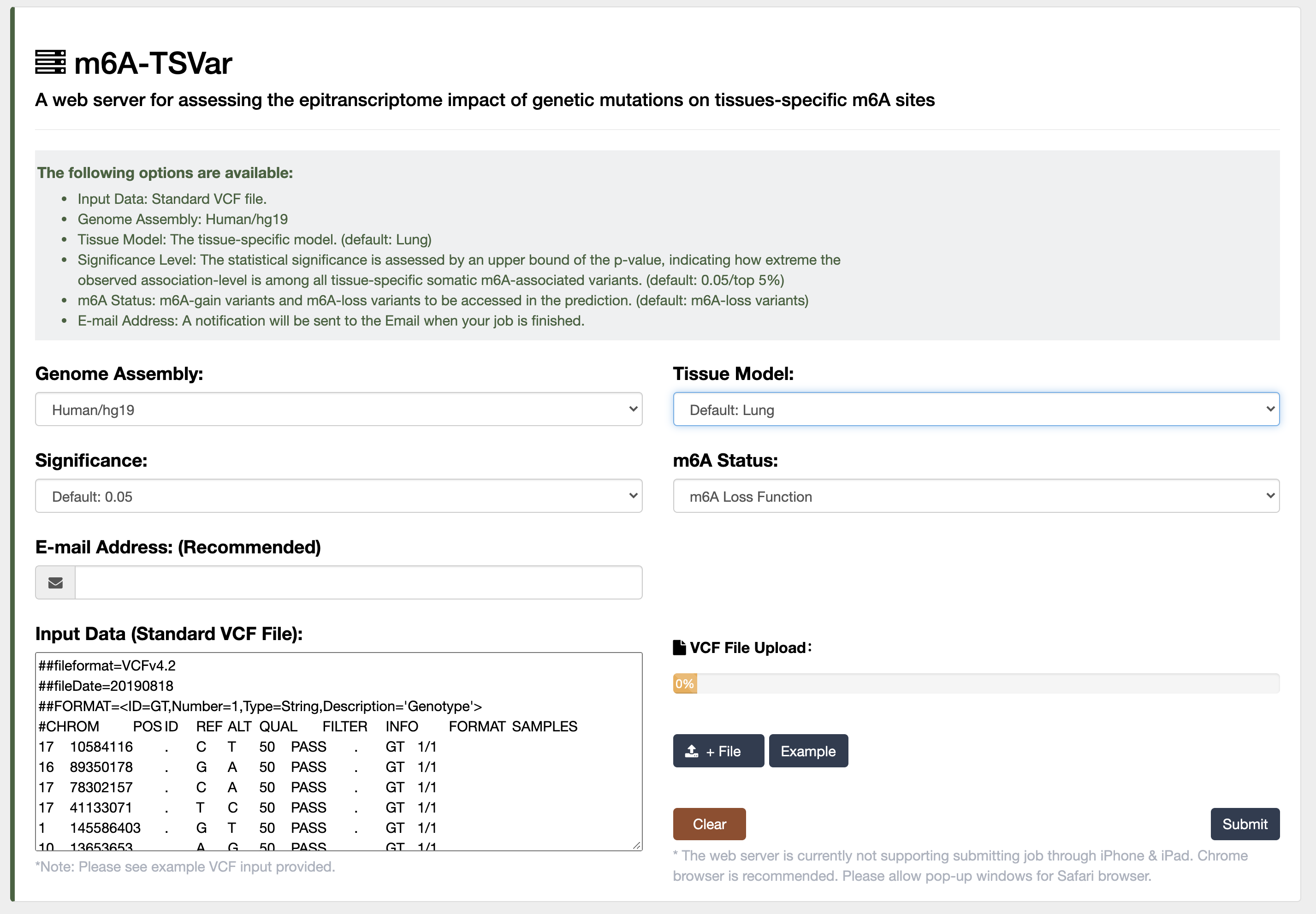
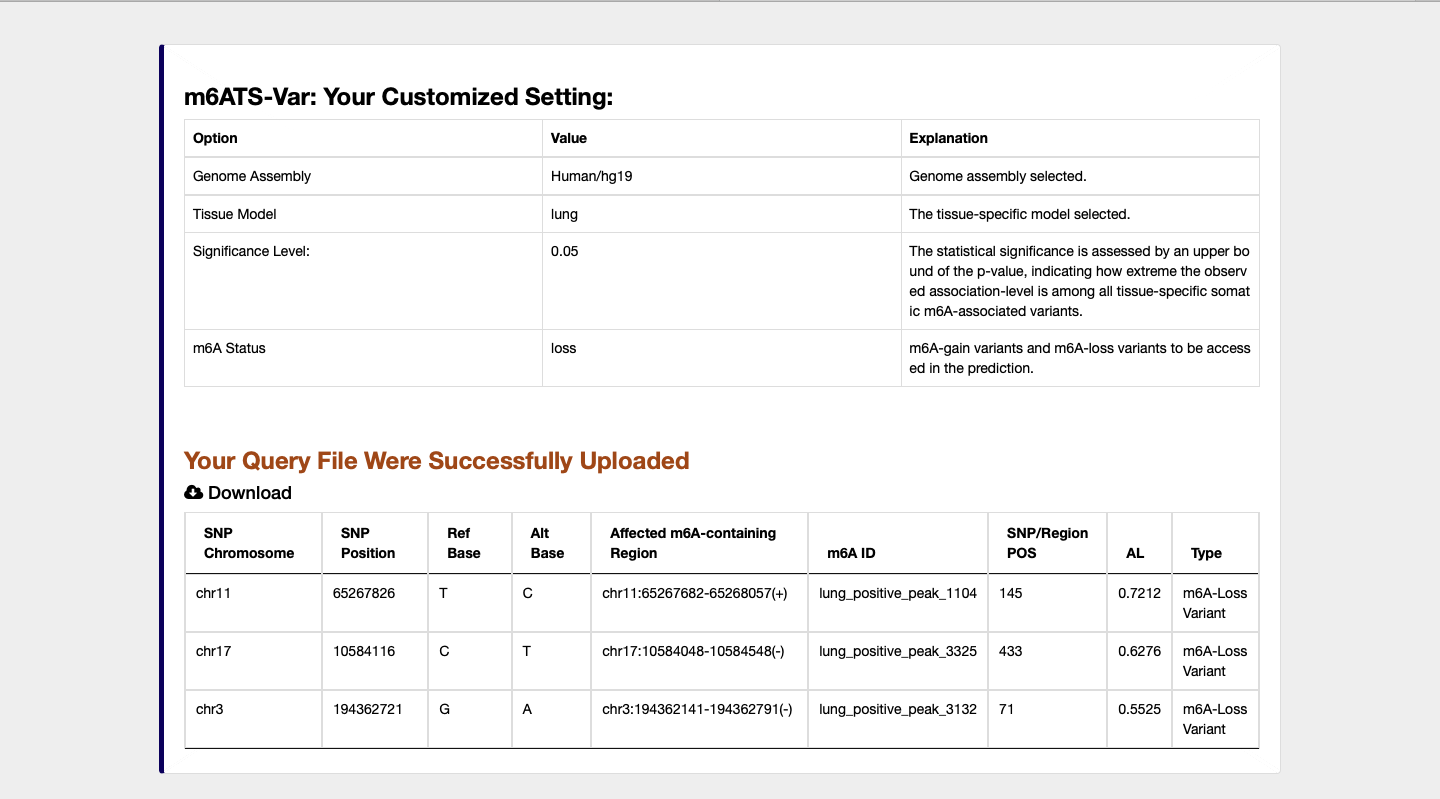
3) m6A-TSVar: a web server for systemically assessing the tissue-specific impact of genetic variants on m6A RNA modification in 23 human tissues.
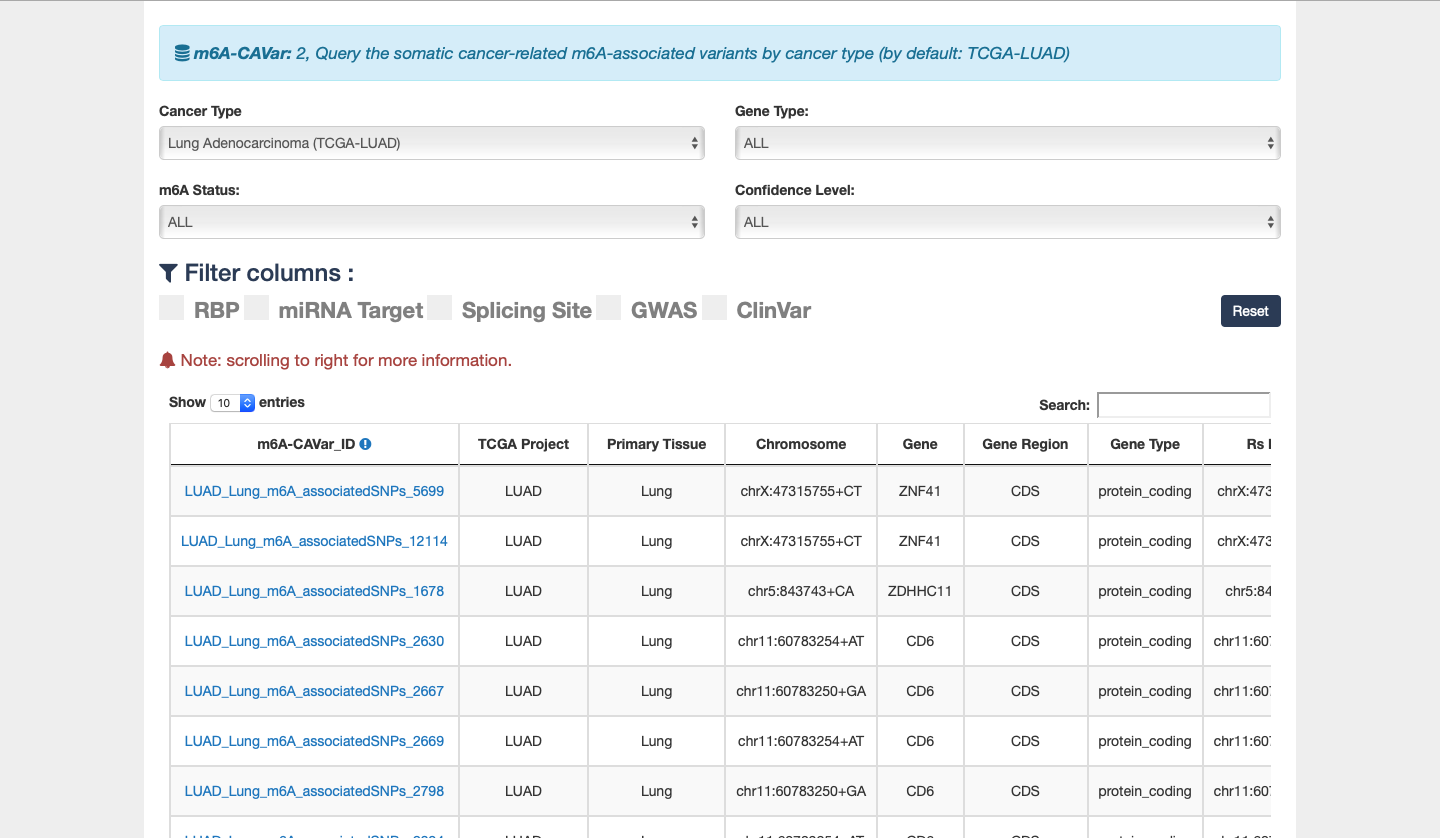
4) m6A-CAVar: a database of 587,983 TCGA cancer mutations (derived from 27 cancer types) that may lead to the gain or loss of m6A sites in the corresponding cancer originating tissues.